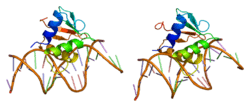SPI1
Transcription factor PU.1 is a protein that in humans is encoded by the SPI1 gene.[5]
Function
This gene encodes an ETS-domain transcription factor that activates gene expression during myeloid and B-lymphoid cell development. The nuclear protein binds to a purine-rich sequence known as the PU-box found near the promoters of target genes, and regulates their expression in coordination with other transcription factors and cofactors. The protein can also regulate alternative splicing of target genes. Multiple transcript variants encoding different isoforms have been found for this gene.[6]
Interactions
SPI1 has been shown to interact with:
See also
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000066336 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000002111 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Ray D, Culine S, Tavitain A, Moreau-Gachelin F (July 1990). "The human homologue of the putative proto-oncogene Spi-1: characterization and expression in tumors". Oncogene. 5 (5): 663–8. PMID 1693183.
- ↑ "Entrez Gene: SPI1 spleen focus forming virus (SFFV) proviral integration oncogene spi1".
- ↑ Hallier M, Lerga A, Barnache S, Tavitian A, Moreau-Gachelin F (February 1998). "The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS". J. Biol. Chem. 273 (9): 4838–42. doi:10.1074/jbc.273.9.4838. PMID 9478924.
- ↑ Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z (July 1999). "Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1". Proc. Natl. Acad. Sci. U.S.A. 96 (15): 8705–10. doi:10.1073/pnas.96.15.8705. PMC 17580. PMID 10411939.
- ↑ Brass AL, Zhu AQ, Singh H (February 1999). "Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers". EMBO J. 18 (4): 977–91. doi:10.1093/emboj/18.4.977. PMC 1171190. PMID 10022840.
- ↑ Escalante CR, Shen L, Escalante MC, Brass AL, Edwards TA, Singh H, Aggarwal AK (July 2002). "Crystallization and characterization of PU.1/IRF-4/DNA ternary complex". J. Struct. Biol. 139 (1): 55–9. doi:10.1016/s1047-8477(02)00514-2. PMID 12372320.
- ↑ Hallier M, Tavitian A, Moreau-Gachelin F (May 1996). "The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb". J. Biol. Chem. 271 (19): 11177–81. doi:10.1074/jbc.271.19.11177. PMID 8626664.
Further reading
- Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA (1990). "The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene". Cell. 61 (1): 113–24. doi:10.1016/0092-8674(90)90219-5. PMID 2180582.
- Nguyen VC, Ray D, Gross MS, de Tand MF, Frézal J, Moreau-Gachelin F (1990). "Localization of the human oncogene SPI1 on chromosome 11, region p11.22". Hum. Genet. 84 (6): 542–6. doi:10.1007/bf00210807. PMID 2338340.
- Chen H, Ray-Gallet D, Zhang P, Hetherington CJ, Gonzalez DA, Zhang DE, Moreau-Gachelin F, Tenen DG (1995). "PU.1 (Spi-1) autoregulates its expression in myeloid cells". Oncogene. 11 (8): 1549–60. PMID 7478579.
- Nagulapalli S, Pongubala JM, Atchison ML (1995). "Multiple proteins physically interact with PU.1. Transcriptional synergy with NF-IL6 beta (C/EBP delta, CRP3)". J. Immunol. 155 (9): 4330–8. PMID 7594592.
- Pongubala JM, Atchison ML (1995). "Activating transcription factor 1 and cyclic AMP response element modulator can modulate the activity of the immunoglobulin kappa 3' enhancer". J. Biol. Chem. 270 (17): 10304–13. doi:10.1074/jbc.270.17.10304. PMID 7730336.
- Hromas R, Orazi A, Neiman RS, Maki R, Van Beveran C, Moore J, Klemsz M (1993). "Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1". Blood. 82 (10): 2998–3004. PMID 8219191.
- Hagemeier C, Bannister AJ, Cook A, Kouzarides T (1993). "The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB". Proc. Natl. Acad. Sci. U.S.A. 90 (4): 1580–4. doi:10.1073/pnas.90.4.1580. PMC 45918. PMID 8434021.
- Hallier M, Tavitian A, Moreau-Gachelin F (1996). "The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb". J. Biol. Chem. 271 (19): 11177–81. doi:10.1074/jbc.271.19.11177. PMID 8626664.
- Bassuk AG, Anandappa RT, Leiden JM (1997). "Physical interactions between Ets and NF-kappaB/NFAT proteins play an important role in their cooperative activation of the human immunodeficiency virus enhancer in T cells". J. Virol. 71 (5): 3563–73. PMC 191503. PMID 9094628.
- Li SL, Valente AJ, Zhao SJ, Clark RA (1997). "PU.1 is essential for p47(phox) promoter activity in myeloid cells". J. Biol. Chem. 272 (28): 17802–9. doi:10.1074/jbc.272.28.17802. PMID 9211934.
- Tsukada J, Misago M, Serino Y, Ogawa R, Murakami S, Nakanishi M, Tonai S, Kominato Y, Morimoto I, Auron PE, Eto S (1997). "Human T-cell leukemia virus type I Tax transactivates the promoter of human prointerleukin-1beta gene through association with two transcription factors, nuclear factor-interleukin-6 and Spi-1". Blood. 90 (8): 3142–53. PMID 9376596.
- Hallier M, Lerga A, Barnache S, Tavitian A, Moreau-Gachelin F (1998). "The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS". J. Biol. Chem. 273 (9): 4838–42. doi:10.1074/jbc.273.9.4838. PMID 9478924.
- Suzuki S, Kumatori A, Haagen IA, Fujii Y, Sadat MA, Jun HL, Tsuji Y, Roos D, Nakamura M (1998). "PU.1 as an essential activator for the expression of gp91(phox) gene in human peripheral neutrophils, monocytes, and B lymphocytes". Proc. Natl. Acad. Sci. U.S.A. 95 (11): 6085–90. doi:10.1073/pnas.95.11.6085. PMC 27589. PMID 9600921.
- Carrère S, Verger A, Flourens A, Stehelin D, Duterque-Coquillaud M (1998). "Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains". Oncogene. 16 (25): 3261–8. doi:10.1038/sj.onc.1201868. PMID 9681824.
- Sato M, Morii E, Takebayashi-Suzuki K, Yasui N, Ochi T, Kitamura Y, Nomura S (1999). "Microphthalmia-associated transcription factor interacts with PU.1 and c-Fos: determination of their subcellular localization". Biochem. Biophys. Res. Commun. 254 (2): 384–7. doi:10.1006/bbrc.1998.9918. PMID 9918847.
- Brass AL, Zhu AQ, Singh H (1999). "Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers". EMBO J. 18 (4): 977–91. doi:10.1093/emboj/18.4.977. PMC 1171190. PMID 10022840.
- Yamamoto H, Kihara-Negishi F, Yamada T, Hashimoto Y, Oikawa T (1999). "Physical and functional interactions between the transcription factor PU.1 and the coactivator CBP". Oncogene. 18 (7): 1495–501. doi:10.1038/sj.onc.1202427. PMID 10050886.
- Rao S, Matsumura A, Yoon J, Simon MC (1999). "SPI-B activates transcription via a unique proline, serine, and threonine domain and exhibits DNA binding affinity differences from PU.1". J. Biol. Chem. 274 (16): 11115–24. doi:10.1074/jbc.274.16.11115. PMID 10196196.
- Mao S, Frank RC, Zhang J, Miyazaki Y, Nimer SD (1999). "Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias". Mol. Cell. Biol. 19 (5): 3635–44. doi:10.1128/mcb.19.5.3635. PMC 84165. PMID 10207087.
External links
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.