TOX
Thymocyte selection-associated high mobility group box protein TOX is a protein that in humans is encoded by the TOX gene.[5][6][7]
Structure and function
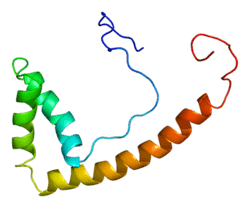
The TOX gene encodes a protein that belongs to a large superfamily of chromatin associated proteins that share an approximately 75 amino acid DNA binding motif, the HMG (high mobility group)-box (named after that found in the canonical member of the family, high mobility group protein 1). Some high mobility group (HMG) box proteins (e.g., LEF1) contain a single HMG box motif and bind DNA in a sequence-specific manner, while other members of this family (e.g., HMGB1) have multiple HMG boxes and bind DNA in a sequence-independent but structure-dependent manner. While TOX has a single HMG-box motif,[7] it is predicted to bind DNA in a sequence-independent manner.[8] TOX is also a member of a small subfamily of proteins (TOX2, TOX3, and TOX4) that share almost identical HMG-box sequences.[8] TOX3 has been identified as a breast cancer susceptibility locus.[9][10] TOX is highly expressed in the thymus, the site of development of T lymphocytes. Knockout mice that lack TOX have a severe defect in development of certain subsets of T lymphocytes.[11]
References
- GRCh38: Ensembl release 89: ENSG00000198846 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000041272 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (Apr 1999). "Prediction of the coding sequences of unidentified human genes. XI. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Res. 5 (5): 277–86. doi:10.1093/dnares/5.5.277. PMID 9872452.
- Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, Kaye J (Mar 2002). "TOX: an HMG box protein implicated in the regulation of thymocyte selection". Nat Immunol. 3 (3): 272–80. doi:10.1038/ni767. PMID 11850626.
- "Entrez Gene: thymocyte selection-associated high mobility group box gene TOX".
- O'Flaherty E, Kaye J (April 2003). "TOX defines a conserved subfamily of HMG-box proteins". BMC Genomics. 4 (1): 13. doi:10.1186/1471-2164-4-13. PMC 155677. PMID 12697058.
- Easton DF, Pooley KA, Dunning AM, et al. (June 2007). "Genome-wide association study identifies novel breast cancer susceptibility loci". Nature. 447 (7148): 1087–93. Bibcode:2007Natur.447.1087E. doi:10.1038/nature05887. PMC 2714974. PMID 17529967.
- Stacey SN, Manolescu A, Sulem P, et al. (July 2007). "Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer". Nat. Genet. 39 (7): 865–9. doi:10.1038/ng2064. PMID 17529974.
- Aliahmad, Parinaz; Kaye, Jonathan (2008-01-21). "Development of all CD4 T lineages requires nuclear factor TOX". The Journal of Experimental Medicine. 205 (1): 245–256. doi:10.1084/jem.20071944. ISSN 1540-9538. PMC 2234360. PMID 18195075.
- Alfei, Francesca; Kanev, Kristiyan; Hofmann, Maike; Wu, Ming; Ghoneim, Hazem E.; Roelli, Patrick; Utzschneider, Daniel T.; von Hoesslin, Madlaina; Cullen, Jolie G. (2019). "TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection". Nature. 571 (7764): 265–269. doi:10.1038/s41586-019-1326-9. ISSN 1476-4687. PMID 31207605.
- Khan, Omar; Giles, Josephine R.; McDonald, Sierra; Manne, Sasikanth; Ngiow, Shin Foong; Patel, Kunal P.; Werner, Michael T.; Huang, Alexander C.; Alexander, Katherine A. (2019). "TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion". Nature. 571 (7764): 211–218. doi:10.1038/s41586-019-1325-x. ISSN 1476-4687. PMC 6713202. PMID 31207603.
- Scott, Andrew C.; Dündar, Friederike; Zumbo, Paul; Chandran, Smita S.; Klebanoff, Christopher A.; Shakiba, Mojdeh; Trivedi, Prerak; Menocal, Laura; Appleby, Heather (2019). "TOX is a critical regulator of tumour-specific T cell differentiation". Nature. 571 (7764): 270–274. doi:10.1038/s41586-019-1324-y. ISSN 1476-4687. PMID 31207604.
Further reading
- Nakajima D, Okazaki N, Yamakawa H, et al. (2003). "Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones". DNA Res. 9 (3): 99–106. doi:10.1093/dnares/9.3.99. PMID 12168954.
- Sebastiani P, Wang L, Nolan VG, et al. (2008). "Fetal hemoglobin in sickle cell anemia: Bayesian modeling of genetic associations". Am. J. Hematol. 83 (3): 189–95. doi:10.1002/ajh.21048. PMID 17918249.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Aliahmad P, O'Flaherty E, Han P, et al. (2004). "TOX provides a link between calcineurin activation and CD8 lineage commitment". J. Exp. Med. 199 (8): 1089–99. doi:10.1084/jem.20040051. PMC 2211890. PMID 15078895.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.