KCNK9
Potassium channel subfamily K member 9 is a protein that in humans is encoded by the KCNK9 gene.[5][6][7]
This gene encodes K2P9.1, one of the members of the superfamily of potassium channel proteins containing two pore-forming P domains. This open channel is highly expressed in the cerebellum. It is inhibited by extracellular acidification and arachidonic acid, and strongly inhibited by phorbol 12-myristate 13-acetate.[7] Phorbol 12-myristate 13-acetate is also known as 12-O-tetradecanoylphorbol-13-acetate (TPA). TASK channels are additionally inhibited by hormones and transmitters that signal through GqPCRs. The resulting cellular depolarization is thought to regulate processes such as motor control and aldosterone secretion. Despite early controversy about the exact mechanism underlying this inhibition, the current view is that Diacyl-glycerol, produced by the breakdown of Phosphatidylinositol-4,5-bis-phosphate by Phospholipase Cβ causes channel closure. [8]
Expression
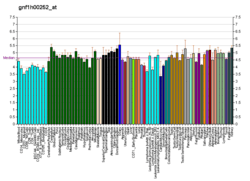
The KCNK9 gene is expressed as an ion channel more commonly known as TASK 3. This channel has a varied pattern of expression. TASK 3 is coexpressed with TASK 1 (KCNK3) in the cerebellar granule cells, locus coeruleus, motor neurons, pontine nuclei, some cells in the neocortex, habenula, olfactory bulb granule cells, and cells in the external plexiform layer of the olfactory bulb.[9] TASK-3 channels are also expressed in the hippocampus; both on pyramidal cells and interneurons.[10] It is thought that these channels may form heterodimers where their expressions co-localise.[11][12]
Function
Mice in which the TASK-3 gene has been deleted have reduced sensitivity to inhalation anaesthetics, exaggerated nocturnal activity and cognitive deficits as well as significantly increased appetite and weight gain.[13][14] A role for TASK-3 channels in neuronal network oscillations has also been described: TASK-3 knockout mice lack the atropine-sensitive halothane-induced theta oscillation (4–7 Hz) from the hippocampus and are unable to maintain theta oscillations during rapid eye movement (REM) sleep.[14]
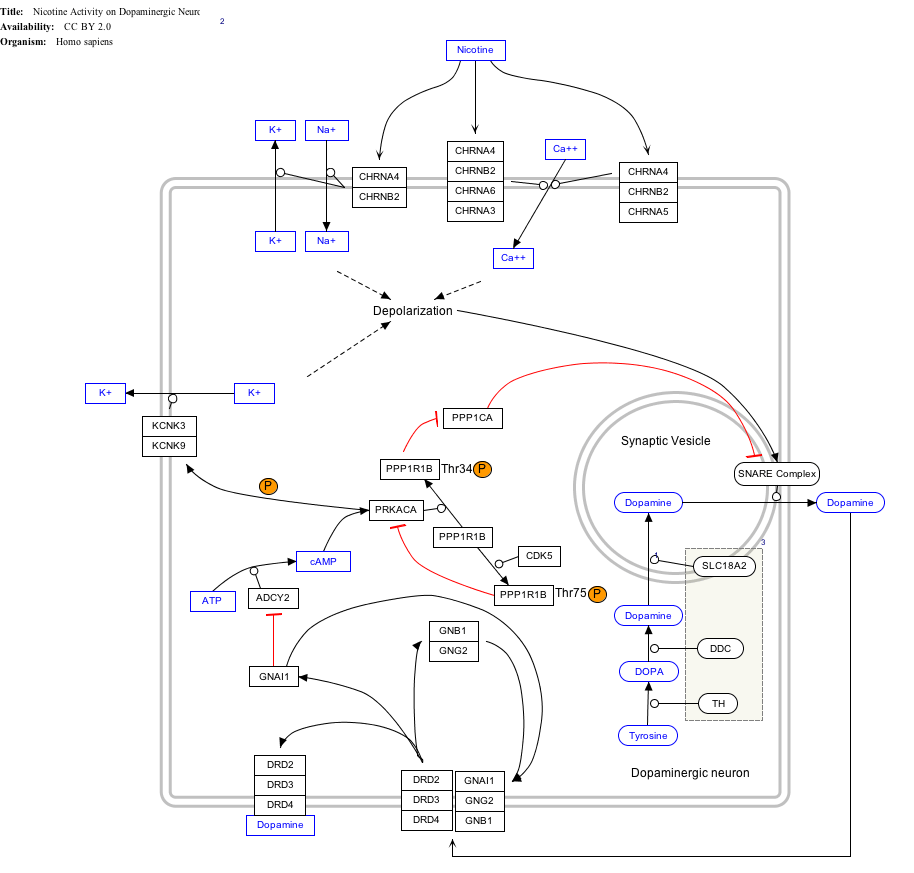
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- ↑ The interactive pathway map can be edited at WikiPathways: "NicotineDopaminergic_WP1602".
See also
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000169427 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000036760 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Kim Y, Bang H, Kim D (May 2000). "TASK-3, a new member of the tandem pore K(+) channel family". J Biol Chem. 275 (13): 9340–7. doi:10.1074/jbc.275.13.9340. PMID 10734076.
- ↑ Goldstein SA, Bayliss DA, Kim D, Lesage F, Plant LD, Rajan S (Dec 2005). "International Union of Pharmacology. LV. Nomenclature and molecular relationships of two-P potassium channels". Pharmacol Rev. 57 (4): 527–40. doi:10.1124/pr.57.4.12. PMID 16382106.
- 1 2 "Entrez Gene: KCNK9 potassium channel, subfamily K, member 9".
- ↑ Wilke, Bettina U.; Lindner, Moritz; Greifenberg, Lea; Albus, Alexandra; Kronimus, Yannick; Bünemann, Moritz; Leitner, Michael G.; Oliver, Dominik (2014-11-25). "Diacylglycerol mediates regulation of TASK potassium channels by Gq-coupled receptors". Nature Communications. 5 (1). doi:10.1038/ncomms6540. ISSN 2041-1723.
- ↑ Bayliss DA, Sirois JE, Talley EM (June 2003). "The TASK family: two-pore domain background K+ channels". Molecular interventions. 3 (4): 205–19. doi:10.1124/mi.3.4.205. PMID 14993448.
- ↑ Torborg CL, Berg AP, Jeffries BW, Bayliss DA, McBain CJ (Jul 12, 2006). "TASK-like conductances are present within hippocampal CA1 stratum oriens interneuron subpopulations". The Journal of Neuroscience. 26 (28): 7362–7. doi:10.1523/jneurosci.1257-06.2006. PMID 16837582.
- ↑ Berg AP, Talley EM, Manger JP, Bayliss DA (Jul 28, 2004). "Motoneurons express heteromeric TWIK-related acid-sensitive K+ (TASK) channels containing TASK-1 (KCNK3) and TASK-3 (KCNK9) subunits". The Journal of Neuroscience. 24 (30): 6693–702. doi:10.1523/jneurosci.1408-04.2004. PMID 15282272.
- ↑ Kang D, Han J, Talley EM, Bayliss DA, Kim D (Jan 1, 2004). "Functional expression of TASK-1/TASK-3 heteromers in cerebellar granule cells". The Journal of Physiology. 554 (Pt 1): 64–77. doi:10.1113/jphysiol.2003.054387. PMC 1664745. PMID 14678492.
- ↑ Linden AM, Aller MI, Leppä E, Rosenberg PH, Wisden W, Korpi ER (October 2008). "K+ channel TASK-1 knockout mice show enhanced sensitivities to ataxic and hypnotic effects of GABA(A) receptor ligands". The Journal of Pharmacology and Experimental Therapeutics. 327 (1): 277–86. doi:10.1124/jpet.108.142083. PMID 18660435.
- 1 2 Pang DS, Robledo CJ, Carr DR, Gent TC, Vyssotski AL, Caley A, Zecharia AY, Wisden W, Brickley SG, Franks NP (Oct 13, 2009). "An unexpected role for TASK-3 potassium channels in network oscillations with implications for sleep mechanisms and anesthetic action". Proceedings of the National Academy of Sciences of the United States of America. 106 (41): 17546–51. doi:10.1073/pnas.0907228106. PMC 2751655. PMID 19805135.
Further reading
- Goldstein SA, Bockenhauer D, O'Kelly I, Zilberberg N (2001). "Potassium leak channels and the KCNK family of two-P-domain subunits". Nat. Rev. Neurosci. 2 (3): 175–84. doi:10.1038/35058574. PMID 11256078.
- Rajan S, Wischmeyer E, Xin Liu G, Preisig-Müller R, Daut J, Karschin A, Derst C (2000). "TASK-3, a novel tandem pore domain acid-sensitive K+ channel. An extracellular histiding as pH sensor". J. Biol. Chem. 275 (22): 16650–7. doi:10.1074/jbc.M000030200. PMID 10747866.
- Chapman CG, Meadows HJ, Godden RJ, Campbell DA, Duckworth M, Kelsell RE, Murdock PR, Randall AD, Rennie GI, Gloger IS (2001). "Cloning, localisation and functional expression of a novel human, cerebellum specific, two pore domain potassium channel". Brain Res. Mol. Brain Res. 82 (1–2): 74–83. doi:10.1016/S0169-328X(00)00183-2. PMID 11042359.
- Vega-Saenz de Miera E, Lau DH, Zhadina M, Pountney D, Coetzee WA, Rudy B (2001). "KT3.2 and KT3.3, two novel human two-pore K(+) channels closely related to TASK-1". J. Neurophysiol. 86 (1): 130–42. PMID 11431495.
- Talley EM, Bayliss DA (2002). "Modulation of TASK-1 (Kcnk3) and TASK-3 (Kcnk9) potassium channels: volatile anesthetics and neurotransmitters share a molecular site of action". J. Biol. Chem. 277 (20): 17733–42. doi:10.1074/jbc.M200502200. PMID 11886861.
- Rajan S, Preisig-Müller R, Wischmeyer E, Nehring R, Hanley PJ, Renigunta V, Musset B, Schlichthörl G, Derst C, Karschin A, Daut J (2003). "Interaction with 14-3-3 proteins promotes functional expression of the potassium channels TASK-1 and TASK-3". J. Physiol. 545 (Pt 1): 13–26. doi:10.1113/jphysiol.2002.027052. PMC 2290646. PMID 12433946.
- Mu D, Chen L, Zhang X, See LH, Koch CM, Yen C, Tong JJ, Spiegel L, Nguyen KC, Servoss A, Peng Y, Pei L, Marks JR, Lowe S, Hoey T, Jan LY, McCombie WR, Wigler MH, Powers S (2003). "Genomic amplification and oncogenic properties of the KCNK9 potassium channel gene". Cancer Cell. 3 (3): 297–302. doi:10.1016/S1535-6108(03)00054-0. PMID 12676587.
- Pei L, Wiser O, Slavin A, Mu D, Powers S, Jan LY, Hoey T (2003). "Oncogenic potential of TASK3 (Kcnk9) depends on K+ channel function". Proc. Natl. Acad. Sci. U.S.A. 100 (13): 7803–7. doi:10.1073/pnas.1232448100. PMC 164668. PMID 12782791.
- Rusznák Z, Pocsai K, Kovács I, Pór A, Pál B, Bíró T, Szücs G (2004). "Differential distribution of TASK-1, TASK-2 and TASK-3 immunoreactivities in the rat and human cerebellum". Cell. Mol. Life Sci. 61 (12): 1532–42. doi:10.1007/s00018-004-4082-3. PMID 15197476.
- Clarke CE, Veale EL, Green PJ, Meadows HJ, Mathie A (2005). "Selective block of the human 2-P domain potassium channel, TASK-3, and the native leak potassium current, IKSO, by zinc". J. Physiol. 560 (Pt 1): 51–62. doi:10.1113/jphysiol.2004.070292. PMC 1665210. PMID 15284350.
- Kim CJ, Cho YG, Jeong SW, Kim YS, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS (2005). "Altered expression of KCNK9 in colorectal cancers". APMIS. 112 (9): 588–94. doi:10.1111/j.1600-0463.2004.apm1120905.x. PMID 15601307.
- Pocsai K, Kosztka L, Bakondi G, Gönczi M, Fodor J, Dienes B, Szentesi P, Kovács I, Feniger-Barish R, Kopf E, Zharhary D, Szucs G, Csernoch L, Rusznák Z (2006). "Melanoma cells exhibit strong intracellular TASK-3-specific immunopositivity in both tissue sections and cell culture". Cell. Mol. Life Sci. 63 (19–20): 2364–76. doi:10.1007/s00018-006-6166-8. PMID 17013562.
- Zuzarte M, Rinné S, Schlichthörl G, Schubert A, Daut J, Preisig-Müller R (2007). "A di-acidic sequence motif enhances the surface expression of the potassium channel TASK-3". Traffic. 8 (8): 1093–100. doi:10.1111/j.1600-0854.2007.00593.x. PMID 17547699.
External links
- KCNK9+protein,+human at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.