MRGPRX4
| MRGPRX4 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | MRGPRX4, GPCR, MRGX4, SNSR6, MAS related GPR family member X4 | ||||||||||||||||||||||||
| External IDs | MGI: 3033139 HomoloGene: 86184 GeneCards: MRGPRX4 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
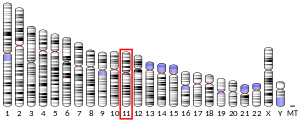
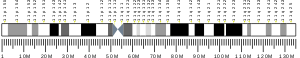
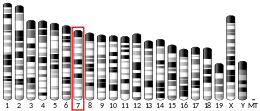
| Location (UCSC) | Chr 11: 18.17 – 18.17 Mb | Chr 7: 48.02 – 48.03 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Mas-related G-protein coupled receptor member X4 is a protein that in humans is encoded by the MRGPRX4 gene.[5][6]
See also
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000179817 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000070552 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ (Sep 2001). "A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons". Cell. 106 (5): 619–32. doi:10.1016/S0092-8674(01)00483-4. PMID 11551509.
- ↑ "Entrez Gene: MRGPRX4 MAS-related GPR, member X4".
Further reading
- Lembo PM, Grazzini E, Groblewski T, et al. (2002). "Proenkephalin A gene products activate a new family of sensory neuron--specific GPCRs". Nat. Neurosci. 5 (3): 201–9. doi:10.1038/nn815. PMID 11850634.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.