Haplogroup M (mtDNA)
| Haplogroup M | |
|---|---|
 | |
| Possible time of origin | 60,000 years before present |
| Possible place of origin | South Asia[1][2][3][4][5][6]later Southeast Asia[7] [8] |
| Ancestor | L3 |
| Descendants | M1, M2, M3, M4'45, M5, M6, M7, M8, M9, M10'42, M12'G, M13, M14, M15, M21, M27, M28, M29'Q, M31'32, M33, M34, M35, M36, M39, M40, M41, M44, M46, M47'50, M48, M49, M51, D |
| Defining mutations | 263, 489, 10400, 14783, 15043[9] |
Haplogroup M is a human mitochondrial DNA (mtDNA) haplogroup. An enormous haplogroup spanning all the continents, the macro-haplogroup M, like its sibling the macro-haplogroup N, is a descendant of the haplogroup L3.
All mtDNA haplogroups considered native outside of Africa are descendants of either haplogroup M or its sibling haplogroup N.[10] Haplogroup M is relatively young, having a younger most recent common ancestor date than some subclades of haplogroup N such as haplogroup R.[11]
Origins
There is a debate concerning geographical origins of Haplogroup M and its sibling haplogroup N. Both lineages are thought to have been the main surviving lineages involved in the out of Africa migration (or migrations) because all indigenous lineages found outside Africa belong to haplogroup M or haplogroup N. Yet to be conclusively determined is whether the mutations that define haplogroups M and N occurred in Africa before the exit from Africa or in Asia after the exit from Africa. Determining the origins of haplogroup M is further complicated by the fact that it is found in Africa and outside of Africa.[3]
It is generally accepted that haplogroup M evolved shortly after the emergence of its parent clade haplogroup L3. Apart from haplogroup M and its sibling haplogroup N, the numerous other subclades of L3 are largely restricted to Africa, which suggests that L3 arose in Africa.
Haplogroup M1
Much of discussion concerning the origins of haplogroup M has been related to its subclade haplogroup M1, which is the only variant of macrohaplogroup M found in Africa.[10] Two possibilities were being considered as potential explanations for the presence of M1 in Africa:
Haplogroup M23
In 2009, two independent publications reported a rare, deep-rooted subclade of haplogroup M, referred to as M23, that is present in Madagascar.[13][14]
The contemporary populations of Madagascar were formed in the last 2,000 years by the admixture of Bantu and Indonesian (Austronesian) populations. M23 seems to be restricted to Madagascar, as it has not been detected anywhere else. M23 could have been brought to Madagascar from Asia where most deep rooted subclades of Haplogroup M are found.
Asian origin hypothesis
According to this theory, anatomically modern humans carrying ancestral haplogroup L3 lineages were involved in the Out of Africa migration from East Africa into Asia. Somewhere in Asia, the ancestral L3 lineages gave rise to haplogroups M and N. The ancestral L3 lineages were then lost by genetic drift as they are infrequent outside Africa. The hypothesis of Asia as the place of origin of macrohaplogroup M is supported by the following:
- The highest frequencies worldwide of macrohaplogroup M are observed in Asia, specifically in Bangladesh, China, India, Japan, Nepal, and Tibet, where frequencies range from 60%-80%. The total frequency of M subclades is even higher in some populations of Siberia or the Americas, but these small populations tend to exhibit strong genetic drift effects, and often their geographical neighbors exhibit very different frequencies.[1][15][16]
- Deep time depth >50,000 years of western, central, southern and eastern Indian haplogroups M2, M38, M54, M58, M33, M6, M61, M62 and the distribution of macrohaplogroup M, do not rule out the possibility of macrohaplogroup M arising in Indian population.[17]
- With the exception of the African specific M1, India has several M lineages that emerged directly from the root of haplogroup M.[1][16]
- Only two subclades of haplogroup M, M1 and M23, are found in Africa, whereas numerous subclades are found outside Africa[1][3] (with some discussion possible only about sub-clade M1, concerning which see below).
- Specifically concerning M1
- Haplogroup M1 has a restricted geographic distribution in Africa, being found mainly in North Africans and East Africa at low or moderate frequencies. If M had originated in Africa around before the Out of Africa migration, it would be expected to have a more widespread distribution [16]
- According to Gonzalez et al. 2007, M1 appears to have expanded relatively recently. In this study M1 had a younger coalescence age than the Asian-exclusive M lineages.[3]
- The geographic distribution of M1 in Africa is predominantly North African/supra-equatorial[3] and is largely confined to Afro-Asiatic speakers,[18] which is inconsistent with the Sub-Saharan distribution of sub-clades of haplogroups L3 and L2 that have similar time depths.[10]
- One of the basal lineages of M1 lineages has been found in Northwest Africa and in the Near East but is absent in East Africa.[3]
- M1 is not restricted to Africa. It is relatively common in the Mediterranean, peaking in Iberia. M1 also enjoys a well-established presence in the Middle East, from the South of the Arabian Peninsula to Anatolia and from the Levant to Iran. In addition, M1 haplotypes have occasionally been observed in the Caucasus and the Trans Caucasus, and without any accompanying L lineages.[3][10] M1 has also been detected in Central Asia, seemingly reaching as far as Tibet.[3]
- The fact that the M1 sub-clade of macrohaplogroup M has a coalescence age which overlaps with that of haplogroup U6 (a Eurasian haplogroup whose presence in Africa is due to a back-migration from West Asia) and the distribution of U6 in Africa is also restricted to the same North African and Horn African populations as M1 supports the scenario that M1 and U6 were part of the same population expansion from Asia to Africa.[18]
- The timing of the proposed migration of M1 and U6-carrying peoples from West Asia to Africa (between 40,000 to 45,000 ybp) is also supported by the fact that it coincides with changes in climatic conditions that reduced the desert areas of North Africa, thereby rendering the region more accessible to entry from the levant. This climatic change also temporally overlaps with the peopling of Europe by populations bearing haplogroup U5, the European sister clade of haplogroup U6.[18]
African origin hypothesis
According to this theory, haplogroups M and N arose from L3 in an East African population that had been isolated from other African populations. Members of this population were involved in the out Africa migration and only carried M and N lineages. With the possible exception of haplogroup M1, all other M and N clades in Africa were lost by genetic drift.[6][12]
The African origin of Haplogroup M is supported by the following arguments and evidence.
- L3, the parent clade of haplogroup M, is found throughout Africa, but is rare outside Africa.[12] According to Toomas Kivisild (2003), "the lack of L3 lineages other than M and N in India and among non-African mitochondria in general suggests that the earliest migration(s) of modern humans already carried these two mtDNA ancestors, via a departure route over the Horn of Africa."[6]
- Specifically concerning at least M1:
- Haplogroup M1 is largely restricted to Africa where the highest frequencies of M1 can be found in Northeast Africa, particularly in Ethiopia. M1 is found in Europe and the Near East but at considerably lower frequencies than in Africa.[3]
Dispersal
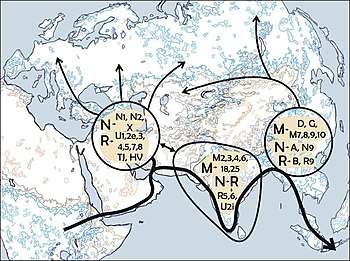
A number of studies have proposed that the ancestors of modern haplogroup M dispersed from Africa through the southern route across the Horn of Africa along the coastal regions of Asia onwards to New Guinea and Australia. These studies suggested that the migrations of haplogroups M and N occurred separately with haplogroup N heading northwards from East Africa to the Levant. However, the results of numerous recent studies indicate that there was only one migration out of Africa and that haplogroups M and N were part of the same migration. This is based on the analysis of a number of relict populations along the proposed beachcombing route from Africa to Australia, all of which possessed both haplogroups N and M.[2][19]
A 2008 study by Abu-Amero et al., suggests that the Arabian Peninsula may have been the main route out of Africa. However, as the region lacks of autochthonous clades of haplogroups M and N the authors suggest that the area has been a more recent receptor of human migrations than an ancient demographic expansion center along the southern coastal route as proposed under the single migration Out-of-Africa scenario of the African origin hypothesis.[4]
Distribution
M is the single most common mtDNA haplogroup in Asia,[20] and peaks in Japan and Tibet, where it represents on average about 70% of the maternal lineages (160/216 = 74% Tibet,[21] 205/282 = 73% Tōkai,[22] 231/326 = 71% Okinawa,[22] 148/211 = 70% Japanese,[15] 50/72 = 69% Tibet,[21] 150/217 = 69% Hokkaidō,[23] 24/35 = 69% Zhongdian Tibetan, 175/256 = 68% northern Kyūshū,[22] 38/56 = 68% Qinghai Tibetan, 16/24 = 67% Diqing Tibetan, 66/100 = 66% Miyazaki, 33/51 = 65% Ainu, 214/336 = 64% Tōhoku,[22] 75/118 = 64% Tokyo (JPT)[24]) and is ubiquitous in India[1][10][25] and South Korea,[22][26][27][28][29] where it has approximately 60% frequency. Among Chinese people both inside and outside of China, haplogroup M accounts for approximately 50% of all mtDNA on average, but the frequency varies from approximately 40% in Hans from Hunan and Fujian in southern China to approximately 60% in Shenyang, Liaoning in northeastern China.[21][22][24][27]
Haplogroup M accounts for approximately 42% of all mtDNA in Filipinos, among whom it is represented mainly by M7c3c and E.[30] In Vietnam, haplogroup M has been found in 37% (52/139) to 48% (20/42) of samples of Vietnamese and in 32% (54/168) of a sample of Chams from Bình Thuận Province.[27][31] Haplogroup M accounts for 43% (92/214) of all mtDNA in a sample of Laotians, with its subclade M7 (M7b, M7c, and M7e) alone accounting for a full third of all haplogroup M, or 14.5% (31/214) of the total sample.[32]
In Oceania, Haplogroup M has been found in 35% (17/48) of a sample of Papua New Guinea highlanders from the Bundi area and in 28% (9/32) of a sample of Aboriginal Australians from Kalumburu in northwestern Australia.[33] A study published in 2008 found Haplogroup M in 42% (60/144) of a pool of samples from nine language groups in the Admiralty Islands of Papua New Guinea, of which 50 belonged to typically Near Oceanian subclades (Q1, Q2) and 10 belonged to typically East or Southeast Asian subclades (M7b, M7c1c, E1b).[34] In a study published in 2015, Haplogroup M was found in 21% (18/86) of a sample of Fijians and in 0% (0/21) of a sample of Rotumans.[35]
Haplogroup M is also relatively common in Northeast Africa, occurring especially among Somalis, Libyans and Oromos at frequencies over 20%.[36][37] Toward the northwest, the lineage is found at comparable frequencies among the Tuareg in Mali and Burkina Faso; particularly the M1a2 subclade (18.42%).[38]
Due to its great age, haplogroup M is an mtDNA lineage which does not correspond well to present-day ethnic groups. It is found among Siberian, Native American, East Asian, Southeast Asian, Central Asian, South Asian, Melanesian, European, Northeast African, and various Middle Eastern populations at varying frequencies.
Among the descendant lineages of haplogroup M are C, D, E, G, Q, and Z. Z and G are found in North Eurasian populations, C and D exists among North Eurasian and Native American populations, E is observed in Southeast Asian populations, and Q is common among Melanesian populations. The lineages M2, M3, M4, M5, M6, M18 and M25 are exclusive to South Asia, with M2 reported to be the oldest lineage on the Indian sub-continent.[1]
In 2013, four ancient specimens dated to around 2,500 BC-500 AD, which were excavated from the Tell Ashara (Terqa) and Tell Masaikh (Kar-Assurnasirpal) archaeological sites in the Euphrates Valley, were found to belong to mtDNA haplotypes associated with the M4b1, M49 and/or M61 haplogroups. Since these clades are not found among the current inhabitants of the area, they are believed to have been brought at a more remote period from east of Mesopotamia; possibly by either merchants or the founders of the ancient Terqa population.[39]
In 2016, three Late Pleistocene European hunter-gatherers were also found to carry M lineages. Two of the specimens were from the Goyet archaeological site in Belgium and were dated to 34,000 and 35,000 years ago, respectively. The other ancient individual hailed from the La Rochette site in France, and was dated to 28,000 years ago.[40]
Ancient DNA analysis of Iberomaurusian skeletal remains at the Taforalt site in Morocco, which have been dated to between 15,100 and 13,900 ybp, observed the M1b subclade among one of the fossils (1/7; ~14%).[41] Ancient individuals belonging to the Late Iron Age settlement of Çemialo Sırtı in Batman, southeast Turkey were found to carry haplogroup M; specifically the M1a1 subclade (1/12; ~8.3%). Haplogroup M was also detected in ancient specimens from Southeast Anatolia (0.4%).[42] Additionally, M1 has been observed among ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from the Pre-Ptolemaic/late New Kingdom and Roman periods.[43] Fossils at the Early Neolithic site of Ifri n'Amr or Moussa in Morocco, which have been dated to around 5,000 BCE, have also been found to carry the M1 subclade. These ancient individuals bore an autochthonous Maghrebi genomic component that peaks among modern Berbers, indicating that they were ancestral to populations in the area.[44] The ancient Egyptian aristocrats Nakht-Ankh and Khnum-Nakht were also found to belong to the M1a1 subclade. The half-brothers lived during the 12th Dynasty, with their tomb located at the Deir Rifeh cemetery in Middle Egypt.[45]
Subgroups distribution
_subclades.png)
- Haplogroup M1'20'51 (14110)
- Haplogroup M1 - found in the Nile Valley, Horn of Africa, Maghreb, Sahara, Mediterranean, and Middle East[1][3]
- M20 - in China.[46]
- M51 - in Cambodia.[47]
- Haplogroup M2 - found in South Asia, with highest concentrations in SE India and Bangladesh;[10] oldest haplogroup M lineage on the Indian sub-continent.[1] Also found with low frequency in southwestern China.[21]
- M2a - most common in Bangladesh
- M2b - most common in SE India
- Haplogroup M3 - found mainly in South Asia, with highest concentrations in west and NW India[10]
- M4'30
- Haplogroup M4 - found mainly in South Asia but some sequences in Eastern Saudi Arabia
- Haplogroup M30 - mainly in India, found in Middle East and North Africa.
- Haplogroup M18'38
- Haplogroup M18 - found among Tharus in southern Nepal and tribal people in Andhra Pradesh[48]
- Haplogroup M38 - found with high frequency among Tharus from Morang District of southeastern Nepal and as singletons among Tharus from Chitwan District of south-central Nepal and Hindus from New Delhi[48]
- Haplogroup M18'38
- Haplogroup M37
- Haplogroup M37a - found in Gujarat, India[16]
- Haplogroup M5 - found in South Asia
- Haplogroup M5a - found in Orissa, India[16]
- Haplogroup M6 - found mainly in South Asia, with highest concentrations in mid-eastern India and Kashmir[10]
- Haplogroup M7 - found in East Asia, especially in Japan, southern China, Vietnam,[49] and Laos[32]
- Haplogroup M8
- Haplogroup M8a: - found in East Asia and Central Asia with low frequency
- Haplogroup CZ
- Haplogroup C - found especially in Siberia
- Haplogroup C1 - found in Asia and America (Native Americans and Hispanics in particular)
- Haplogroup C4
- Haplogroup C7 - found in China and Ukraine.
- Haplogroup Z - found in Northeast Europe, Central Asia and East Asia, including among Finns, Hazara, Japanese, Koreans, Chinese, Russians, and Sami
- Haplogroup C - found especially in Siberia
- Haplogroup M9 - found in East Asia and Central Asia, especially in Tibet
- Haplogroup E - a subclade of M9 - found especially in Taiwan (aborigines), Maritime Southeast Asia, and the Mariana Islands
- Haplogroup M10 - small clade found in East Asia, Southeast Asia, Bangladesh, Central Asia, southern Siberia, and Belarus
- Haplogroup M11 - small clade found especially among the Chinese and also in some Japanese, Koreans, Oroqen, Yi, Tibetans, and Bangladeshis[21]
- Haplogroup M12'G
- Haplogroup M12 - small clade found especially among the aborigines of Hainan Island as well as in other populations of China,[24] Japan, Korea, Pashtuns, Tibet, and Vietnam
- Haplogroup G - found especially in Japan, Mongolia, and Tibet and in indigenous peoples of Kamchatka (Koryaks, Alyutors, Itelmens), with some isolated instances in diverse places of Asia
- Haplogroup M13 - small clade found among Tibetans in Tibet,[21] Oirat Mongols in Xinjiang,[51] Barghuts in Hulunbuir,[52] and Yakuts and Dolgans in central Siberia[53]
- Haplogroup M14 - found in Tibet[21]
- Haplogroup M15 - found in Tibet[21]
- Haplogroup M17 - found in Luzon,[30] Chams,[31][54] Maniq,[54] Mon,[54] Blang,[54] Lawa,[54] Thai,[54] and Laotians[54]
- Haplogroup M19 - found in the Batak people of Palawan[55]
- Haplogroup M21 - small clade found in SE Asia (Semang, Semelai, Temuan,[54] Jehai,[54] Thailand, Maniq,[54] Mon,[54] Karen[54]) and Bangladesh
- Haplogroup M23'75
- M23 - found in Madagascar
- M75 - found in China[46]
- Haplogroup M24 - found in Palawan[55]
- Haplogroup M27 - found in Melanesia
- Haplogroup M28 - found in Melanesia and in a single Han individual from Taiwan[21]
- Haplogroup M29'Q
- Haplogroup M29 - found in Melanesia
- Haplogroup Q - found in Melanesia and Australia (Aborigines)
- Haplogroup M31 - found among the Onge, in the Andaman Islands[16]
- Haplogroup M32 - found in Andaman Islands
- Haplogroup M33 - small clade found in South Asia, Belarus, southern China,[24] and in two Han Chinese living in Southern California[21]
- Haplogroup M33a - found in Gujarat, India[16]
- Haplogroup M34 - small clade found in South Asia
- Haplogroup M35 - small clade found in South Asia
- Haplogroup M35a - found in India
- Haplogroup M35b - found in Karnataka, India and Nepal. Found in Slovakia.[56]
- Haplogroup M39 - found in South Asia[16]
- Haplogroup M40 - found in South Asia[16]
- Haplogroup M41 - found in South Asia
- Haplogroup M41b - found in Andhra Pradesh, India[16]
- Haplogroup M41c - found in Andhra Pradesh, India[16]
- Haplogroup M42 - found among Australian Aborigines
- Haplogroup M48 - rare clade found in Saudi Arabia
- Haplogroup M49 - found among ancient specimens in the Euphrates valley
- Haplogroup M80 - found in Batak people of Palawan[55]
- Haplogroup D - found in Eastern Eurasia, Native Americans, Central Asia[57] and occasionally also in West Asia and Europe.
Subclades
Tree
This phylogenetic tree of haplogroup M subclades is based on the paper by Mannis van Oven and Manfred Kayser Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation[9] and subsequent published research.
- M
- M1
- M1a
- M1a1
- M1a1a
- M1a1b
- M1a1b1
- M1a1c
- M1a1d
- M1a1e
- M1a1f
- M1a2
- M1a2a
- M1a2b
- M1a3
- M1a3a
- M1a3b
- M1a4
- M1a5
- M1a1
- M1b
- M1b1
- M1b1a
- M1b2
- M1b2a
- M1b1
- M1a
- M2
- M2a
- M2a1
- M2a2
- M2a3
- M2b
- M2b1
- M2b2
- M2a
- M3
- M3a
- M4"45
- M4
- M4a
- M4b
- M4b1
- M18'38
- M18
- M38
- M30
- M30a
- M30b
- M30c
- M30c1
- M30c1a
- M30c1a1
- M30c1a
- M30c1
- M30d
- M37
- M37a
- M43
- M45
- M4
- M5
- M5a
- M5a1
- M5a1a
- M5a1b
- M5a2
- M5a2a
- M5a1
- M5a
- M6
- M7
- M7a
- M7a1
- M7a1a
- M7a1a1
- M7a1a1a
- M7a1a2
- M7a1a3
- M7a1a4
- M7a1a4a
- M7a1a5
- M7a1a6
- M7a1a7
- M7a1a1
- M7a1b
- M7a1a
- M7a2
- M7a2a
- M7a2b
- M7a1
- M7b'c'd'e
- M7b'd
- M7b
- M7b1'2
- M7b1
- M7b2
- M7b2a
- M7b2b
- M7b2c
- M7b3
- M7b3a
- M7b1'2
- M7d
- M7b
- M7c'e
- M7c
- M7c1
- M7c1a
- M7c1b
- M7c1b1
- M7c2
- M7c2a
- M7c3
- M7c3a
- M7c3b
- M7c3c
- M7c1
- M7e
- M7c
- M7b'd
- M7a
- M8
- M9
- M9a'b'c'd
- M9a'c'd
- M9a'd
- M9a
- M9a1
- M9a2
- M9a3
- M9d
- M9a
- M9c
- M9a'd
- M9b
- M9a'c'd
- E
- M9a'b'c'd
- M10'42
- M10
- M10a
- M10a1
- M10a2
- M10a
- M42
- M42a
- M10
- M11
- M11a
- M11b
- M12'G
- M12
- M12a
- G
- M12
- M13
- M13a
- M13a1
- M13a
- M14
- M15
- M21
- M21a'b
- M21a
- M21b
- M21c'd
- M21c
- M21d
- M21a'b
- M22
- M23
- M25
- M27
- M27a
- M27b
- M27c
- M28
- M28a
- M28b
- M29'Q
- M29
- M29a
- M29b
- Q
- M29
- M31'32
- M31
- M31a
- M31a1
- M31a1a
- M31a1b
- M31a2
- M31a2a
- M31a1
- M31b
- M31c
- M31a
- M32
- M32a
- M31
- M33
- M33a
- M33b
- M33c
- M34
- M34a
- M35
- M35a
- M35b
- M36
- M36a
- M39
- M39a
- M40
- M40a
- M41
- M44'52
- M44
- M52
- M46
- M47'50
- M47
- M50
- M48
- M49
- M51
- D
- M1
See also
| Wikimedia Commons has media related to Haplogroup M (mtDNA). |
- Genealogical DNA test
- Genetic Genealogy
- Human mitochondrial genetics
- Population Genetics
- Human mitochondrial DNA haplogroups
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||
References
- 1 2 3 4 5 6 7 8 Rajkumar; et al. (2005). "Phylogeny and antiquity of M macrohaplogroup inferred from complete mt DNA sequence of Indian specific lineages". BMC Evolutionary Biology. 5: 26. doi:10.1186/1471-2148-5-26. PMC 1079809. PMID 15804362.
- 1 2 Macaulay, V; Hill, C; Achilli, A; Rengo, C; Clarke, D; Meehan, W; Blackburn, J; Semino, O; et al. (2005). "Single, Rapid Coastal Settlement of Asia Revealed by Analysis of Complete Mitochondrial Genomes". Science. 308 (5724): 1034–6. doi:10.1126/science.1109792. PMID 15890885. : "Haplogroup L3 (the African clade that gave rise to the two basal non-African clades, haplogroups M and N) is 84,000 years old, and haplogroups M and N themselves are almost identical in age at 63,000 years old, with haplogroup R diverging rapidly within haplogroup N 60,000 years ago."
- 1 2 3 4 5 6 7 8 9 10 11 Gonzalez; et al. (2007). "Mitochondrial lineage M1 traces an early human backflow to Africa". BMC Genomics. 8: 223. doi:10.1186/1471-2164-8-223. PMC 1945034. PMID 17620140.
- 1 2 Abu-Amero, KK; Larruga, JM; Cabrera, VM; González, AM; et al. (2008). "Mitochondrial DNA structure in the Arabian Peninsula". BMC Evolutionary Biology. BMC Evolutionary Biology. 8: 45. doi:10.1186/1471-2148-8-45. PMC 2268671. PMID 18269758.
- ↑ Kivisild, M; Kivisild, T; Metspalu, E; Parik, J; Hudjashov, G; Kaldma, K; Serk, P; Karmin, M; Behar, DM; Gilbert, M Thomas P; Endicott, Phillip; Mastana, Sarabjit; Papiha, Surinder S; Skorecki, Karl; Torroni, Antonio; Villems, Richard (2004). "Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans". BMC Genetics. 5: 26. doi:10.1186/1471-2156-5-26. PMC 516768. PMID 15339343.
- 1 2 3 Kivisild, T; Rootsi, S; Metspalu, M; Mastana, S; Kaldma, K; Parik, J; Metspalu, E; Adojaan, M; et al. (2003). "The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations". American Journal of Human Genetics. 72 (2): 313–32. doi:10.1086/346068. PMC 379225. PMID 12536373.
- ↑ https://bmcevolbiol.biomedcentral.com/articles/10.1186/s12862-016-0816-8
- ↑ Quintana-Murci, Lluís; et al. (1999). "Where West Meets East: The Complex mtDNA Landscape of the Southwest and Central Asian Corridor". Am. J. Hum. Genet. 74: 828. doi:10.1086/383236.
- 1 2 van Oven, M; Kayser, M; et al. (2009). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation. 30 (2): E386–E394. doi:10.1002/humu.20921. PMID 18853457.
- 1 2 3 4 5 6 7 8 Metspalu, M; Kivisild, T; Metspalu, E; Parik, J; Hudjashov, G; Kaldma, K; Serk, P; Karmin, M; Behar, DM; Gilbert, M Thomas P; Endicott, Phillip; Mastana, Sarabjit; Papiha, Surinder S; Skorecki, Karl; Torroni, Antonio; Villems, Richard; et al. (2004). "Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans". BMC Genetics. 5: 26. doi:10.1186/1471-2156-5-26. PMC 516768. PMID 15339343.
- ↑ Fregel, R; Cabrera, V; Larruga, JM; Abu-Amero, KK; González, AM (2015). "Carriers of Mitochondrial DNA Macrohaplogroup N Lineages Reached Australia around 50,000 Years Ago following a Northern Asian Route". PLOS ONE. 10: e0129839. doi:10.1371/journal.pone.0129839. PMC 4460043. PMID 26053380.
- 1 2 3 Quintana; et al. (1999). "Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa" (PDF).
- ↑ Dubut, V; Cartault, F; Payet, C; Thionville, MD; Murail, P; et al. (2009). "Complete mitochondrial sequences for haplogroups M23 and M46: insights into the Asian ancestry of the Malagasy population". Human Biology. 81 (4): 495–500. doi:10.3378/027.081.0407. PMID 20067372.
- ↑ Ricaut, FX; Razafindrazaka, H; Cox, MP; Dugoujon, JM; Guitard, E; Sambo, C; Mormina, M; Mirazon-Lahr, M; Ludes, B; Crubézy, Eric; et al. (2009). "A new deep branch of eurasian mtDNA macrohaplogroup M reveals additional complexity regarding the settlement of Madagascar". BMC Genomics. 10: 605. doi:10.1186/1471-2164-10-605. PMC 2808327. PMID 20003445.
- 1 2 Maruyama, Sayaka; Minaguchi, Kiyoshi; Saitou, Naruya (2003). "Sequence polymorphisms of the mitochondrial DNA control region and phylogenetic analysis of mtDNA lineages in the Japanese population". Int J Legal Med. 117: 218–225. doi:10.1007/s00414-003-0379-2.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Thangaraj et al. (2006), In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup 'M' in India, BMC Genomics 2006, 7:151
- ↑ Chandrasekar Adimoolam, Kumar S; Sreenath, J; Sarkar, BN; Urade, BP; et al. (2009). "Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern Human in South Asian Corridor". PLoS ONE. 4 (10): e7447. doi:10.1371/journal.pone.0007447. PMC 2757894. PMID 19823670.
- 1 2 3 Olivieri et al. (2006), The mtDNA legacy of the Levantine early Upper Palaeolithic in Africa, Science. 2006 Dec 15;314(5806):1767-70
- ↑ Hudjashov, Kivisild, G; Kivisild, T; Underhill, PA; Endicott, P; Sanchez, JJ; Lin, AA; Shen, P; Oefner, P; et al. (2007). "Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis". Proceedings of the National Academy of Sciences of the United States of America. 104 (21): 8726–30. doi:10.1073/pnas.0702928104. PMC 1885570. PMID 17496137.
- ↑ Ghezzi; et al. (2005). "Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson's disease in Italians". European Journal of Human Genetics. 13: 748–752. doi:10.1038/sj.ejhg.5201425. PMID 15827561.
- 1 2 3 4 5 6 7 8 9 10 Ji, Fuyun; Sharpley, Mark S.; Derbeneva, Olga; et al. (2012). "Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans". PNAS. 109 (19): 7391–7396. doi:10.1073/pnas.1202484109. PMC 3358837. PMID 22517755.
- 1 2 3 4 5 6 Umetsu, Kazuo; Tanaka, Masashi; Yuasa, Isao; et al. (2005). "Multiplex amplified product-length polymorphism analysis of 36 mitochondrial single-nucleotide polymorphisms for haplogrouping of East Asian populations". Electrophoresis. 26: 91–98. doi:10.1002/elps.200406129.
- ↑ Asari, Masaru; Umetsu, Kazuo; Adachi, Noboru; et al. (2007). "", "Utility of haplogroup determination for forensic mtDNA analysis in the Japanese population". Legal Medicine. 9: 237–240. doi:10.1016/j.legalmed.2007.01.007.
- 1 2 3 4 Zheng, H-X; Yan, S; Qin, Z-D; Wang, Y; Tan, J-Z; et al. (2011). "Major Population Expansion of East Asians Began before Neolithic Time: Evidence of mtDNA Genomes". PLoS ONE. 6 (10): e25835. doi:10.1371/journal.pone.0025835. PMC 3188578. PMID 21998705.
- ↑ Edwin; et al. (2002). "Mitochondrial DNA diversity among five tribal populations of southern India" (PDF). Current Science. 83.
- ↑ Kim, W; Yoo, T-K; Shin, D-J; Rho, H-W; Jin, H-J; et al. (2008). "Mitochondrial DNA Haplogroup Analysis Reveals no Association between the Common Genetic Lineages and Prostate Cancer in the Korean Population". PLoS ONE. 3 (5): e2211. doi:10.1371/journal.pone.0002211. PMC 2376063. PMID 18493608.
- 1 2 3 Jin, H-J; Tyler-Smith, C; Kim, W (2009). "The Peopling of Korea Revealed by Analyses of Mitochondrial DNA and Y-Chromosomal Markers". PLoS ONE. 4 (1): e4210. doi:10.1371/journal.pone.0004210. PMC 2615218. PMID 19148289.
- 1 2 Derenko, Miroslava; Malyarchuk, Boris; Grzybowski, Tomasz; et al. (2007). "Phylogeographic Analysis of Mitochondrial DNA in Northern Asian Populations". Am. J. Hum. Genet. 81: 1025–1041. doi:10.1086/522933. PMC 2265662. PMID 17924343.
- ↑ Beom Hong, Seung; Cheol Kim, Ki; Kim, Wook (2014). "Mitochondrial DNA haplogroups and homogeneity in the Korean population". Genes & Genomics. 36: 583–590. doi:10.1007/s13258-014-0194-9.
- 1 2 Tabbada, Kristina A.; Trejaut, Jean; Loo, Jun-Hun; et al. (Jan 2010). "Philippine Mitochondrial DNA Diversity: A Populated Viaduct between Taiwan and Indonesia?". Mol. Biol. Evol. 27 (1): 21–31. doi:10.1093/molbev/msp215. PMID 19755666.
- 1 2 Peng, Min-Sheng; Ho Quang, Huy; Pham Dang, Khoa; et al. (2010). "Tracing the Austronesian Footprint in Mainland Southeast Asia: A Perspective from Mitochondrial DNA". Mol. Biol. Evol. 27 (10): 2417–2430. doi:10.1093/molbev/msq131. PMID 20513740.
- 1 2 Bodner, Martin; Zimmermann, Bettina; Röck, Alexander; et al. (2011). "Southeast Asian diversity: first insights into the complex mtDNA structure of Laos". BMC Evolutionary Biology. 11: 49. doi:10.1186/1471-2148-11-49. PMC 3050724. PMID 21333001.
- ↑ Hudjashov, Georgi; Kivisild, Toomas; Underhill, Peter A.; et al. (2007). "Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis". PNAS. 104 (21): 8726–8730. doi:10.1073/pnas.0702928104. PMC 1885570. PMID 17496137.
- ↑ Kayser, Manfred; Choi, Ying; van Oven, Mannis; et al. (July 2008). "", (2008) "The Impact of the Austronesian Expansion: Evidence from mtDNA and Y Chromosome Diversity in the Admiralty Islands of Melanesia". Molecular Biology and Evolution. 25 (7): 1362–1374. doi:10.1093/molbev/msn078. PMID 18390477.
- ↑ Shipley, G. P.; Taylor, D. A.; Tyagi, A.; Tiwari, G.; Redd, A. (2015). "Genetic structure among Fijian island populations". Journal of Human Genetics. 60 (2): 69–75. doi:10.1038/jhg.2014.105.
- ↑ Non, Amy. "ANALYSES OF GENETIC DATA WITHIN AN INTERDISCIPLINARY FRAMEWORK TO INVESTIGATE RECENT HUMAN EVOLUTIONARY HISTORY AND COMPLEX DISEASE" (PDF). University of Florida. Retrieved 12 April 2016.
- ↑ Holden. "MtDNA variation in North, East, and Central African populations gives clues to a possible back-migration from the Middle East". American Association of Physical Anthropologists. Archived from the original on 3 March 2016. Retrieved 13 April 2016.
- ↑ Luísa Pereira; Viktor Černý; María Cerezo; Nuno M Silva; Martin Hájek; Alžběta Vašíková; Martina Kujanová; Radim Brdička; Antonio Salas (17 March 2010). "Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel". European Journal of Human Genetics. 18: 915–923. doi:10.1038/ejhg.2010.21. PMC 2987384. PMID 20234393. Retrieved 20 May 2016.
- ↑ Witas HW, Tomczyk J, Jędrychowska-Dańska K, Chaubey G, Płoszaj T (2013). "mtDNA from the Early Bronze Age to the Roman Period Suggests a Genetic Link between the Indian Subcontinent and Mesopotamian Cradle of Civilization". PLoS ONE. 8 (9): e73682. doi:10.1371/journal.pone.0073682. PMC 3770703. PMID 24040024. Retrieved 20 May 2016.
- ↑ Cosimo Posth; Gabriel Renaud; Alissa Mittnik; Dorothée G. Drucker; Hélène Rougier; Christophe Cupillard; Frédérique Valentin; Corinne Thevenet; Anja Furtwängler; Christoph Wißing; Michael Francken; Maria Malina; Michael Bolus; Martina Lari; Elena Gigli; Giulia Capecchi; Isabelle Crevecoeur; Cédric Beauval; Damien Flas; Mietje Germonpré; Johannes van der Plicht; Richard Cottiaux; Bernard Gély; Annamaria Ronchitelli; Kurt Wehrberger; Dan Grigorescu; Jiří Svoboda; Patrick Semal; David Caramelli; Hervé Bocherens; Katerina Harvati; Nicholas J. Conard; Wolfgang Haak; Adam Powell (March 21, 2016). "Pleistocene Mitochondrial Genomes Suggest a Single Major Dispersal of Non-Africans and a Late Glacial Population Turnover in Europe". Current Biology. 26: 1–7. doi:10.1016/j.cub.2016.01.037. PMID 26853362. Retrieved 5 June 2016.
- ↑ van de Loosdrecht et al. (2018-03-15). "Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations". Science. American Association for the Advancement of Science (AAAS): eaar8380. doi:10.1126/science.aar8380. ISSN 0036-8075.
- ↑ Reyhan Yaka. "Archaeogenetics of Late Iron Age Çemialo Sırtı, Batman: Investigating maternal genetic continuity in North Mesopotamia since the Neolithic". bioRxiv 172890.
- ↑ Schuenemann, Verena J.; et al. (2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications. 8: 15694. doi:10.1038/ncomms15694. PMC 5459999. PMID 28556824.
- ↑ Fregel; et al. (2018). "Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe". bioRxiv 191569.
- ↑ Konstantina Drosou, Campbell Price, Terence A. Brown (February 2018). "The kinship of two 12th Dynasty mummies revealed by ancient DNA sequencing" (PDF). Journal of Archaeological Science. 17: 793–797. doi:10.1016/j.jasrep.2017.12.025.
- 1 2 Kong, Qing-Peng et al 2010, Large-Scale mtDNA Screening Reveals a Surprising Matrilineal Complexity in East Asia and Its Implications to the Peopling of the Region
- ↑ Hartmann et al. 2009, Validation of microarray-based resequencing of 93 worldwide mitochondrial genomes
- 1 2 Fornarino, Simona; Pala, Maria; Battaglia, Vincenza; et al. (2009). "Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation". BMC Evolutionary Biology. 9: 154. doi:10.1186/1471-2148-9-154. PMC 2720951. PMID 19573232.
- ↑ Peng; et al. (2011). "Tracing the legacy of the early Hainan Islanders - a perspective from mitochondrial DNA". BMC Evolutionary Biology. 11: 46. doi:10.1186/1471-2148-11-46. PMC 3048540. PMID 21324107.
- ↑ Tanaka, Masashi; Cabrera, Vicente M.; González, Ana M.; et al. (October 2004). "Mitochondrial Genome Variation in Eastern Asia and the Peopling of Japan". Genome Res. 14 (10): 1832–1850. doi:10.1101/gr.2286304. PMC 524407. PMID 15466285.
- ↑ Yao, Yong-Gang; Kong, Qing-Peng; Wang, Cheng-Ye; et al. (2004). "Different Matrilineal Contributions to Genetic Structure of Ethnic Groups in the Silk Road Region in China". Mol. Biol. Evol. 21 (12): 2265–2280. doi:10.1093/molbev/msh238.
- ↑ Derenko, M; Malyarchuk, B; Denisova, G; Perkova, M; Rogalla, U; et al. (2012). "Complete Mitochondrial DNA Analysis of Eastern Eurasian Haplogroups Rarely Found in Populations of Northern Asia and Eastern Europe". PLoS ONE. 7 (2): e32179. doi:10.1371/journal.pone.0032179. PMC 3283723. PMID 22363811.
- ↑ Fedorova, Sardana A; Reidla, Maere; Metspalu, Ene; et al. (2013). "Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia". BMC Evolutionary Biology. 13: 127. doi:10.1186/1471-2148-13-127. PMC 3695835. PMID 23782551.
- 1 2 3 4 5 6 7 8 9 10 11 12 Kutanan, Wibhu; Kampuansai, Jatupol; Changmai, Piya; et al. (2018). "Contrasting maternal and paternal genetic variation of hunter-gatherer groups in Thailand". Scientific Reports. 8: 1536. doi:10.1038/s41598-018-20020-0.
- 1 2 3 Scholes, Clarissa; Siddle, Katherine; Ducourneau, Axel; et al. (2011). "Genetic Diversity and Evidence for Population Admixture in Batak Negritos from Palawan". American Journal of Physical Anthropology. 146: 62–72. doi:10.1002/ajpa.21544. PMID 21796613.
- ↑ Malyarchuk, B. et al 2008c, Mitochondrial DNA Variability in Slovaks, with Application to the Roma Origin,
- ↑ Comas et al. (2004), Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages, European Journal of Human Genetics (2004) 12, 495–504.
External links
- General
- Ian Logan's Mitochondrial DNA Site
- The India DNA geographical project at Family Tree DNA
- The China DNA geographical project at Family Tree DNA
- Haplogroup M
- Mannis van Oven's PhyloTree.org - mtDNA subtree M
- Spread of Haplogroup M, from National Geographic
- Tree of M haplogroup as for 2006
- Haplogroup M (mtDNA) interest group on Facebook
- Another tree emphasizing the Andamanese and Nicobarese populations in comparison with other peoples with high M presence
- K.Tharanghaj et al. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup M in India (PDF document)