Chromosome 16
| Chromosome 16 | |
|---|---|
 Human chromosome 16 pair after G-banding. One is from mother, one is from father. | |
 Chromosome 16 pair in human male karyogram. | |
| Features | |
| Length (bp) |
90,338,345 bp (GRCh38)[1] |
| No. of genes | 795 (CCDS)[2] |
| Type | Autosome |
| Centromere position |
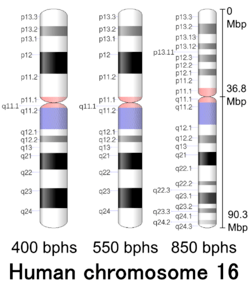
Metacentric[3] (36.8 Mbp[4]) |
| Complete gene lists | |
| CCDS | Gene list |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome 16 |
| Entrez | Chromosome 16 |
| NCBI | Chromosome 16 |
| UCSC | Chromosome 16 |
| Full DNA sequences | |
| RefSeq | NC_000016 (FASTA) |
| GenBank | CM000678 (FASTA) |
Chromosome 16 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 16 spans about 90 million base pairs (the building material of DNA) and represents just under 3% of the total DNA in cells.
Genes
Number of genes
The following are some of the gene count estimates of human chromosome 16. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[5]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 795 | - | - | [2] | 2016-09-08 |
| HGNC | 802 | 251 | 365 | [6] | 2017-05-12 |
| Ensembl | 865 | 1,046 | 462 | [7] | 2017-03-29 |
| UniProt | 838 | - | - | [8] | 2018-02-28 |
| NCBI | 912 | 652 | 502 | [9][10][11] | 2017-05-19 |
Gene list
The following is a partial list of genes on human chromosome 16. For complete list, see the link in the infobox on the right.
- ACSF3: encoding enzyme Acyl-CoA synthetase family member 3
- ACSM2B: encoding enzyme Acyl-coenzyme A synthetase ACSM2B, mitochondrial
- ACSM3: encoding enzyme Acyl-coenzyme A synthetase ACSM3, mitochondrial 2
- ADHD1: Attention deficit-hyperactivity disorder, susceptibility to, 1
- ARL6IP1: encoding protein ADP-ribosylation factor-like protein 6-interacting protein 1
- ARMC5
- BMIQ5: Body mass index quantitative trait locus 5
- C16orf13/JFP2: encoding protein Chromosome 16 open reading frame 13
- C16orf42: encoding [[]] FALSE
- C16orf58: encoding protein Chromosome 16 open reading frame 58
- C16orf62: encoding protein Chromosome 16 open reading frame 62
- C16orf71: encoding protein Uncharacterized protein Chromosome 16 Open Reading Frame 71
- C16orf84:
- C16orf95:
- C16orf96:
- C16orf96: encoding protein C16orf96, or chromosome 16 open reading frame 96,
- CARHSP1: Calcium-regulated heat stable protein 1
- CASP16P: encoding protein Caspase 16, pseudogene
- CCDC113: encoding protein Coiled-coil domain-containing protein 113
- Ccdc78: encoding protein Coiled-coil domain-containing 78 (CCDC78)
- CDIPT: CDP-diacylglycerol-inositol 3-phosphatidyltransferase
- CFDP1: Craniofacial development protein 1
- CHDS1: Coronary heart disease, susceptibility to, 1
- CIAPIN1: Anamorsin (originally, Cytokine induced apoptosis inhibitor 1)
- CKLF: Chemokine-like factor
- CLUAP1:
- CMTM2: encoding protein CKLF-like MARVEL transmembrane domain-containing protein 2
- CCDC135: encoding protein Coiled-coil domain-containing protein 135
- COTL1: encoding protein Coactosin-like protein
- CTRL: Chymotrypsin-like protease
- DCTPP1: encoding enzyme dCTP pyrophosphatase 1
- DEL16p13.3, RSTSS: Chromosome 16p13.3 deletion syndrome (Rubinstein-Taybi deletion syndrome)
- DHX38: DEAH-box helicase 38
- DUP16p13.3, C16DUPq13.3: Chromosome 16p13.3 duplication syndrome
- EMP2: Epithelial membrane protein 2
- ENKD1: Enkurin domain-containing protein 1
- ERAF: Alpha-hemoglobin-stabilizing protein
- FAHD1: Fumarylacetoacetate hydrolase domain-containing protein 1
- FAM57B: Family with sequence similarity 57 member B
- FBRS: Probably fibrosin-1 long transcript protein
- FOXC2-AS1: encoding protein FOXC2 antisense RNA 1
- GLG1: Golgi apparatus protein 1
- HBAP1: Hemoglobin, alpha pseudogene 1
- HBHR, ATR1: Alpha-thalassemia/mental retardation syndrome, type 1
- HIRIP3: encoding protein HIRA-interacting protein 3
- HN1L: encoding protein Hematological and neurological expressed 1-like protein
- IBD8: Inflammatory bowel disease 8
- IHPS2: Pyloric stenosis, infantile hypertrophic, 2
- ITFG3: encoding protein Protein ITFG3
- KDM8: encoding protein Lysine demethylase 8
- LINC00273 encoding protein Long intergenic non-protein coding RNA 273
- LOC124220: encoding protein Zymogen granule protein 16 homolog B
- LOC81691:
- LUC7L: encoding protein Putative RNA-binding protein Luc7-like 1
- LYPLA3: encoding enzyme Group XV phospholipase A2
- MC1R: melanocortin 1 receptor
- MCOPCT1: Microphthalmia with cataract 1
- MT1G: encoding protein Metallothionein-1G
- METRN: encoding protein Meteorin, glial cell differentiation regulator
- MKL2: encoding protein MKL/myocardin-like protein 2
- MPHOSPH6: encoding enzyme M-phase phosphoprotein 6
- MT1X: encoding protein Metallothionein 1X
- NIP30: encoding protein NIP30 protein
- NOB1: encoding protein RNA-binding protein NOB1
- NOMO1: encoding protein Nodal modulator 1
- NPW: encoding protein Neuropeptide W
- NUBP2: encoding protein Nucleotide-binding protein 2
- NUPR1: encoding protein Nuclear protein 1
- OGFOD1:
- PDF: encoding enzyme Peptide deformylase, mitochondrial
- PDPR: encoding protein Pyruvate dehydrogenase phosphatase regulatory subunit
- PKDTS: Polycystic kidney disease, infantile severe, with tuberous sclerosis
- PMFBP1: encoding protein Polyamine-modulated factor 1-binding protein 1
- POLR3K: encoding enzyme DNA-directed RNA polymerase III subunit RPC10
- PRR35: encoding protein Proline rich 35
- RPS15A: encoding protein 40S ribosomal protein S15a
- RSL1D1: encoding protein Ribosomal L1 domain-containing protein 1
- SHCBP1: encoding protein SHC SH2 domain-binding protein 1
- SLZ1: encoding protein SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
- SNAI3-AS1: encoding protein SNAI3 antisense RNA 1
- SNORD71: encoding protein Small nucleolar RNA, C/D box 71
- SPSB3: encoding protein SplA/ryanodine receptor domain and SOCS box containing 3
- SRCAP: encoding enzyme Helicase SRCAP
- TANGO6: encoding protein Transport and Golgi organization protein 6 homolog
- TAO2: encoding Serine/threonine-protein kinase TAO2
- TBC1D24: encoding protein TBC1 domain family, member 24
- TELO2: encoding protein Telomere length regulation protein TEL2 homolog
- TMEM112: encoding enzyme Lipase maturation factor 1
- TMEM8A: encoding protein Transmembrane protein 8A
- TNRC6A: encoding protein Trinucleotide repeat-containing gene 6A protein
- Tuberous sclerosis complex tumor suppressors: encoding [[]] FALSE
- UNKL: encoding protein RING finger protein unkempt-like
- VAT1L: encoding protein Vesicle amine transport protein 1 homolog (T. californica)-like
- ZG16
- ZNF23: encoding protein Zinc finger protein 23
- ZNF200: encoding protein Zinc finger protein 200
- ZNF263: encoding protein Zinc finger protein 263
- ZNF629: encoding protein Zinc finger protein 629
Diseases and disorders
Associated traits
Cytogenetic band
| Chr. | Arm[19] | Band[20] | ISCN start[21] |
ISCN stop[21] |
Basepair start |
Basepair stop |
Stain[22] | Density |
|---|---|---|---|---|---|---|---|---|
| 16 | p | 13.3 | 0 | 352 | 1 | 7,800,000 | gneg | |
| 16 | p | 13.2 | 352 | 596 | 7,800,001 | 10,400,000 | gpos | 50 |
| 16 | p | 13.13 | 596 | 813 | 10,400,001 | 12,500,000 | gneg | |
| 16 | p | 13.12 | 813 | 948 | 12,500,001 | 14,700,000 | gpos | 50 |
| 16 | p | 13.11 | 948 | 1070 | 14,700,001 | 16,700,000 | gneg | |
| 16 | p | 12.3 | 1070 | 1246 | 16,700,001 | 21,200,000 | gpos | 50 |
| 16 | p | 12.2 | 1246 | 1409 | 21,200,001 | 24,200,000 | gneg | |
| 16 | p | 12.1 | 1409 | 1558 | 24,200,001 | 28,500,000 | gpos | 50 |
| 16 | p | 11.2 | 1558 | 1856 | 28,500,001 | 35,300,000 | gneg | |
| 16 | p | 11.1 | 1856 | 2045 | 35,300,001 | 36,800,000 | acen | |
| 16 | q | 11.1 | 2045 | 2194 | 36,800,001 | 38,400,000 | acen | |
| 16 | q | 11.2 | 2194 | 2709 | 38,400,001 | 47,000,000 | gvar | |
| 16 | q | 12.1 | 2709 | 2953 | 47,000,001 | 52,600,000 | gneg | |
| 16 | q | 12.2 | 2953 | 3142 | 52,600,001 | 56,000,000 | gpos | 50 |
| 16 | q | 13 | 3142 | 3346 | 56,000,001 | 57,300,000 | gneg | |
| 16 | q | 21 | 3346 | 3657 | 57,300,001 | 66,600,000 | gpos | 100 |
| 16 | q | 22.1 | 3657 | 4023 | 66,600,001 | 70,800,000 | gneg | |
| 16 | q | 22.2 | 4023 | 4118 | 70,800,001 | 72,800,000 | gpos | 50 |
| 16 | q | 22.3 | 4118 | 4294 | 72,800,001 | 74,100,000 | gneg | |
| 16 | q | 23.1 | 4294 | 4551 | 74,100,001 | 79,200,000 | gpos | 75 |
| 16 | q | 23.2 | 4551 | 4659 | 79,200,001 | 81,600,000 | gneg | |
| 16 | q | 23.3 | 4659 | 4768 | 81,600,001 | 84,100,000 | gpos | 50 |
| 16 | q | 24.1 | 4768 | 4930 | 84,100,001 | 87,000,000 | gneg | |
| 16 | q | 24.2 | 4930 | 5025 | 87,000,001 | 88,700,000 | gpos | 25 |
| 16 | q | 24.3 | 5025 | 5120 | 88,700,001 | 90,338,345 | gneg |
References
- ↑ "Human Genome Assembly GRCh38 - Genome Reference Consortium". National Center for Biotechnology Information. 2013-12-24. Retrieved 2017-03-04.
- 1 2 "Search results - 16[CHR] AND "Homo sapiens"[Organism] AND ("has ccds"[Properties] AND alive[prop]) - Gene". NCBI. CCDS Release 20 for Homo sapiens. 2016-09-08. Retrieved 2017-05-28.
- ↑ Tom Strachan; Andrew Read (2 April 2010). Human Molecular Genetics. Garland Science. p. 45. ISBN 978-1-136-84407-2.
- 1 2 Genome Decoration Page, NCBI. Ideogram data for Homo sapience (850 bphs, Assembly GRCh38.p3). Last update 2014-06-03. Retrieved 2017-04-26.
- ↑ Pertea M, Salzberg SL (2010). "Between a chicken and a grape: estimating the number of human genes". Genome Biol. 11 (5): 206. doi:10.1186/gb-2010-11-5-206. PMC 2898077. PMID 20441615.
- ↑ "Statistics & Downloads for chromosome 16". HUGO Gene Nomenclature Committee. 2017-05-12. Retrieved 2017-05-19.
- ↑ "Chromosome 16: Chromosome summary - Homo sapiens". Ensembl Release 88. 2017-03-29. Retrieved 2017-05-19.
- ↑ "Human chromosome 16: entries, gene names and cross-references to MIM". UniProt. 2018-02-28. Retrieved 2018-03-16.
- ↑ "Search results - 16[CHR] AND "Homo sapiens"[Organism] AND ("genetype protein coding"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ↑ "Search results - 16[CHR] AND "Homo sapiens"[Organism] AND ( ("genetype miscrna"[Properties] OR "genetype ncrna"[Properties] OR "genetype rrna"[Properties] OR "genetype trna"[Properties] OR "genetype scrna"[Properties] OR "genetype snrna"[Properties] OR "genetype snorna"[Properties]) NOT "genetype protein coding"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ↑ "Search results - 16[CHR] AND "Homo sapiens"[Organism] AND ("genetype pseudo"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ↑ Maillard, A M (25 November 2014). "The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity". Molecular Psychiatry. 20: 140–147. doi:10.1038/mp.2014.145. PMID 25421402.
- ↑ Richter, M (21 February 2018). "Altered TAOK2 activity causes autism-related neurodevelopmental and cognitive abnormalities through RhoA signaling". Molecular Psychiatry. doi:10.1038/s41380-018-0025-5. PMID 29467497.
- ↑ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (400 bphs, Assembly GRCh38.p3). Last update 2014-03-04. Retrieved 2017-04-26.
- ↑ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (550 bphs, Assembly GRCh38.p3). Last update 2015-08-11. Retrieved 2017-04-26.
- ↑ International Standing Committee on Human Cytogenetic Nomenclature (2013). ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013). Karger Medical and Scientific Publishers. ISBN 978-3-318-02253-7.
- ↑ Sethakulvichai, W.; Manitpornsut, S.; Wiboonrat, M.; Lilakiatsakun, W.; Assawamakin, A.; Tongsima, S. (2012). "Estimation of band level resolutions of human chromosome images" (PDF). In Computer Science and Software Engineering (JCSSE), 2012 International Joint Conference on: 276–282. doi:10.1109/JCSSE.2012.6261965.
- ↑ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (850 bphs, Assembly GRCh38.p3). Last update 2014-06-03. Retrieved 2017-04-26.
- ↑ "p": Short arm; "q": Long arm.
- ↑ For cytogenetic banding nomenclature, see article locus.
- 1 2 These values (ISCN start/stop) are based on the length of bands/ideograms from the ISCN book, An International System for Human Cytogenetic Nomenclature (2013). Arbitrary unit.
- ↑ gpos: Region which is positively stained by G banding, generally AT-rich and gene poor; gneg: Region which is negatively stained by G banding, generally CG-rich and gene rich; acen Centromere. var: Variable region; stalk: Stalk.
- Gilbert F (1999). "Disease genes and chromosomes: disease maps of the human genome. Chromosome 16". Genet Test. 3 (2): 243–54. PMID 10464676.
- Martin J, et al. (2004). "The sequence and analysis of duplication-rich human chromosome 16". Nature. 432 (7020): 988–94. doi:10.1038/nature03187. PMID 15616553.
- Miller, David T; Nasir, Ramzi; Sobeih, Magdi M; Shen, Yiping; Wu, Bai-Lin; Hanson, Ellen (2011-10-27). 16p11.2 Microdeletion. PMID 20301775. NBK11167. In Pagon RA, Bird TD, Dolan CR, et al., eds. (1993). GeneReviews™ [Internet]. Seattle WA: University of Washington, Seattle.
- http://omim.org/search?index=geneMap&search=16p13.3
External links
| Wikimedia Commons has media related to Human chromosome 16. |
- National Institutes of Health. "Chromosome 16". Genetics Home Reference. Retrieved 2017-05-06.
- "Chromosome 16". Human Genome Project Information Archive 1990–2003. Retrieved 2017-05-06.