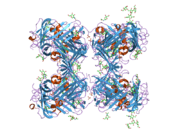Urokinase receptor
The Urokinase receptor, also known as uPA receptor or uPAR or CD87 (Cluster of Differentiation 87), is multidomain glycoprotein tethered to the cell membrane with a glycosylphosphotidylinositol (GPI) anchor. uPAR was originally identified as a saturable binding site for urokinase on the cell surface.
Molecular characteristics
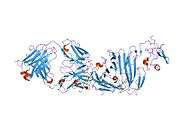
uPAR consists of three different domains of the Ly-6/uPAR/alpha-neurotoxin family. All three domains are necessary for high affinity binding of the primary ligand, urokinase. It has been possible to express uPAR recombinantly in CHO-cells and S2 cells from Drosophila melanogaster. 4 out of 5 of the possible glycosylation sites are used in vivo giving the protein a molecular weight of 50-60 kDA. Recently the structure of uPAR was solved by X-ray crystallography in complex with a peptide antagonist[5] and with its native ligand, urokinase.[6]
Besides the primary ligand urokinase, uPAR interacts with several other proteins, among others: vitronectin, the uPAR associated protein (uPARAP) and the integrin family of membrane proteins.
Physiological significance
uPAR is a part of the plasminogen activation system, which in the healthy body is involved in tissue reorganization events such as mammary gland involution and wound healing. In order to be able to reorganize tissue, the old tissue must be able to be degraded. An important mechanism in this degradation is the proteolysis cascade initiated by the plasminogen activation system. uPAR binds urokinase and thus restricts plasminogen activation to the immediate vicinity of the cell membrane. Thus uPAR seems to be an important player in the regulation of this process.
However the components of the plasminogen activation system have been found to be highly expressed in many malignant tumors, indicating that tumors are able to hijack the system, and use it in metastasis. Thus inhibitors of the various components of the plasminogen activation system have been sought as possible anticancer drugs.
uPAR has been involved in various other non-proteolytical processes related to cancer, such as cell migration, cell cycle regulation, and cell adhesion.
When uPA is bound to the receptor, there is cleavage between the GPI-anchor and the uPAR, releasing suPAR.[7]
Interactions
See also
- Cancer
- Cluster of differentiation
- Metastasis
- Plasmin
- Plasminogen activation system
- Urokinase
- suPAR
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000011422 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000046223 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Llinas P, Le Du MH, Gårdsvoll H, Danø K, Ploug M, Gilquin B, Stura EA, Ménez A (May 2005). "Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide". The EMBO Journal. 24 (9): 1655–63. doi:10.1038/sj.emboj.7600635. PMC 1142576. PMID 15861141.
- ↑ Huai Q, Mazar AP, Kuo A, Parry GC, Shaw DE, Callahan J, Li Y, Yuan C, Bian C, Chen L, Furie B, Furie BC, Cines DB, Huang M (February 2006). "Structure of human urokinase plasminogen activator in complex with its receptor". Science. 311 (5761): 656–9. doi:10.1126/science.1121143. PMID 16456079.
- ↑ Thunø M, Macho B, Eugen-Olsen J (2009). "suPAR: the molecular crystal ball". Disease Markers. 27 (3): 157–72. doi:10.3233/DMA-2009-0657. PMID 19893210.
- ↑ Czekay RP, Kuemmel TA, Orlando RA, Farquhar MG (May 2001). "Direct binding of occupied urokinase receptor (uPAR) to LDL receptor-related protein is required for endocytosis of uPAR and regulation of cell surface urokinase activity". Molecular Biology of the Cell. 12 (5): 1467–79. doi:10.1091/mbc.12.5.1467. PMC 34598. PMID 11359936.
Further reading
- Ploug M (2003). "Structure-function relationships in the interaction between the urokinase-type plasminogen activator and its receptor". Current Pharmaceutical Design. 9 (19): 1499–528. doi:10.2174/1381612033454630. PMID 12871065.
- Kjøller L (January 2002). "The urokinase plasminogen activator receptor in the regulation of the actin cytoskeleton and cell motility". Biological Chemistry. 383 (1): 5–19. doi:10.1515/BC.2002.002. PMID 11928822.
- Chavakis T, Kanse SM, May AE, Preissner KT (April 2002). "Haemostatic factors occupy new territory: the role of the urokinase receptor system and kininogen in inflammation". Biochemical Society Transactions. 30 (2): 168–73. doi:10.1042/BST0300168. PMID 12023845.
- Ploug M, Gårdsvoll H, Jørgensen TJ, Lønborg Hansen L, Danø K (April 2002). "Structural analysis of the interaction between urokinase-type plasminogen activator and its receptor: a potential target for anti-invasive cancer therapy". Biochemical Society Transactions. 30 (2): 177–83. doi:10.1042/BST0300177. PMID 12023847.
- Alfano M, Sidenius N, Blasi F, Poli G (November 2003). "The role of urokinase-type plasminogen activator (uPA)/uPA receptor in HIV-1 infection". Journal of Leukocyte Biology. 74 (5): 750–6. doi:10.1189/jlb.0403176. PMID 12960238.
- Alfano D, Franco P, Vocca I, Gambi N, Pisa V, Mancini A, Caputi M, Carriero MV, Iaccarino I, Stoppelli MP (February 2005). "The urokinase plasminogen activator and its receptor: role in cell growth and apoptosis". Thrombosis and Haemostasis. 93 (2): 205–11. doi:10.1160/TH04-09-0592. PMID 15711734.
External links
- PLAUR+protein,+human at the US National Library of Medicine Medical Subject Headings (MeSH)