Drosophila melanogaster
| Drosophila melanogaster | |
|---|---|
 | |
| Drosophila melanogaster | |
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Arthropoda |
| Class: | Insecta |
| Order: | Diptera |
| Family: | Drosophilidae |
| Genus: | Drosophila |
| Subgenus: | Sophophora |
| Species group: | Drosophila melanogaster group |
| Species subgroup: | Drosophila melanogaster subgroup |
| Species complex: | Drosophila melanogaster complex |
| Species: | D. melanogaster |
| Binomial name | |
| Drosophila melanogaster | |
.jpg)
.jpg)
Drosophila melanogaster is a species of fly (the taxonomic order Diptera) in the family Drosophilidae. The species is known generally as the common fruit fly (though inaccurately[2]) or vinegar fly. Starting with Charles W. Woodworth's proposal of the use of this species as a model organism, D. melanogaster continues to be widely used for biological research in genetics, physiology, microbial pathogenesis, and life history evolution. As of 2017, eight Nobel prizes had been awarded for research using Drosophila.[3]
Drosophila is typically used in research because it can be readily reared in the laboratory, has only four pairs of chromosomes, breeds quickly, and lays many eggs.[4] Its geographic range includes all continents, including islands.[5] D. melanogaster is a common pest in homes, restaurants, and other places where food is served.[6]
Flies belonging to the family Tephritidae are also called "fruit flies". This can cause confusion, especially in the Mediterranean, Australia and South Africa, where the Mediterranean fruit fly Ceratitis capitata is an economic pest.
Lifecycle and reproduction
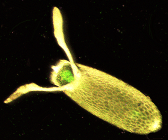
The D. melanogaster lifespan is about 21 days from egg to death.[7]
The developmental period for D. melanogaster varies with temperature, as with many ectothermic species. The shortest development time (egg to adult), 7 days, is achieved at 28 °C (82 °F).[8][9] Development times increase at higher temperatures (11 days at 30 °C or 86 °F) due to heat stress. Under ideal conditions, the development time at 25 °C (77 °F) is 8.5 days,[8][9][10] at 18 °C (64 °F) it takes 19 days[8][9] and at 12 °C (54 °F) it takes over 50 days.[8][9] Under crowded conditions, development time increases,[11] while the emerging flies are smaller.[11][12] Females lay some 400 eggs (embryos), about five at a time, into rotting fruit or other suitable material such as decaying mushrooms and sap fluxes. The eggs, which are about 0.5 mm long, hatch after 12–15 hours (at 25 °C or 77 °F).[8][9] The resulting larvae grow for about 4 days (at 25 °C) while molting twice (into second- and third-instar larvae), at about 24 and 48 h after hatching.[8][9] During this time, they feed on the microorganisms that decompose the fruit, as well as on the sugar of the fruit itself. The mother puts feces on the egg sacs to establish the same microbial composition in the larvae's guts which has worked positively for herself.[13] Then the larvae encapsulate in the puparium and undergo a four-day-long metamorphosis (at 25 °C), after which the adults eclose (emerge).[8][9]
The female fruit fly prefers a shorter duration when it comes to sex. Males, on the other hand, prefer it to last longer.[14] Males perform a sequence of five behavioral patterns to court females. First, males orient themselves while playing a courtship song by horizontally extending and vibrating their wings. Soon after, the male positions itself at the rear of the female's abdomen in a low posture to tap and lick the female genitalia. Finally, the male curls its abdomen and attempts copulation. Females can reject males by moving away, kicking, and extruding their ovipositor.[15] Copulation lasts around 15–20 minutes,[16] during which males transfer a few hundred, very long (1.76 mm) sperm cells in seminal fluid to the female.[17] Females store the sperm in a tubular receptacle and in two mushroom-shaped spermathecae; sperm from multiple matings compete for fertilization. A last male precedence is believed to exist: the last male to mate with a female sires about 80% of her offspring. This precedence was found to occur through both displacement and incapacitation.[18] The displacement is attributed to sperm handling by the female fly as multiple matings are conducted and is most significant during the first 1–2 days after copulation. Displacement from the seminal receptacle is more significant than displacement from the spermathecae.[18] Incapacitation of first male sperm by second male sperm becomes significant 2–7 days after copulation. The seminal fluid of the second male is believed to be responsible for this incapacitation mechanism (without removal of first male sperm) which takes effect before fertilization occurs.[18] The delay in effectiveness of the incapacitation mechanism is believed to be a protective mechanism that prevents a male fly from incapacitating its own sperm should it mate with the same female fly repetitively. Sensory neurons in the uterus of female D. melanogaster respond to a male protein, sex peptide, which is found in sperm.[19] This protein makes the female reluctant to copulate for about 10 days after insemination. The signal pathway leading to this change in behavior has been determined. The signal is sent to a brain region that is a homolog of the hypothalamus and the hypothalamus then controls sexual behavior and desire.[19]
D. melanogaster is often used for life extension studies, such as to identify genes purported to increase lifespan when mutated.[20]
Females

Females become receptive to courting males at about 8–12 hours after emergence.[22] Specific neuron groups in females have been found to affect copulation behavior and mate choice. One such group in the abdominal nerve cord allows the female fly to pause her body movements to copulate.[19] Activation of these neurons induces the female to cease movement and orient herself towards the male to allow for mounting. If the group is inactivated, the female remains in motion and does not copulate. Various chemical signals such as male pheromones often are able to activate the group.[19]
Also, females exhibit mate choice copying. When virgin females are shown other females copulating with a certain type of male, they tend to copulate more with this type of male afterwards than naive females (which have not observed the copulation of others). This behavior is sensitive to environmental conditions, and females copulate less in bad weather conditions.[23]
Males
D. melanogaster males exhibit a strong reproductive learning curve. That is, with sexual experience, these flies tend to modify their future mating behavior in multiple ways. These changes include increased selectivity for courting only intraspecifically, as well as decreased courtship times.
Sexually naïve D. melanogaster males are known to spend significant time courting interspecifically, such as with D. simulans flies. Naïve D. melanogaster will also attempt to court females that are not yet sexually mature, and other males. D. melanogaster males show little to no preference for D. melanogaster females over females of other species or even other male flies. However, after D. simulans or other flies incapable of copulation have rejected the males’ advances, D. melanogaster males are much less likely to spend time courting nonspecifically in the future. This apparent learned behavior modification seems to be evolutionarily significant, as it allows the males to avoid investing energy into futile sexual encounters.[24]
In addition, males with previous sexual experience will modify their courtship dance when attempting to mate with new females — the experienced males spend less time courting and therefore have lower mating latencies, meaning that they are able to reproduce more quickly. This decreased mating latency leads to a greater mating efficiency for experienced males over naïve males.[25] This modification also appears to have obvious evolutionary advantages, as increased mating efficiency is extremely important in the eyes of natural selection.
Polygamy
Both male and female D. melanogaster act polygamously (having multiple sexual partners at the same time).[26] In both males and females, polygamy results in a decrease in evening activity compared to virgin flies, more so in males than females.[26] Evening activity consists of the activities that the flies participate in other than mating and finding partners, such as finding food.[27] The reproductive success of males and females varies, due to the fact that a female only needs to mate once to reach maximum fertility.[27] Mating with multiple partners provides no advantage over mating with one partner, and therefore females exhibit no difference in evening activity between polygamous and monogamous individuals.[27] For males, however, mating with multiple partners increases their reproductive success by increasing the genetic diversity of their offspring.[27] This benefit of genetic diversity is an evolutionary advantage because it increases the chance that some of the offspring will have traits that increase their fitness in their environment.
The difference in evening activity between polygamous and monogamous male flies can be explained with courtship. For polygamous flies, their reproductive success increases by having offspring with multiple partners, and therefore they spend more time and energy on courting multiple females.[27] On the other hand, monogamous flies only court one female, and expend less energy doing so.[27] While it requires more energy for male flies to court multiple females, the overall reproductive benefits it produces has kept polygamy as the preferred sexual choice.[27]
It has been shown that the mechanism that affects courtship behavior in Drosophila is controlled by the oscillator neurons DN1s and LNDs.[28] Oscillation of the DN1 neurons was found to be effected by socio-sexual interactions, and is connected to mating-related decrease of evening activity.[28]
Physical appearance

Wildtype fruit flies are yellow-brown, with brick-red eyes and transverse black rings across the abdomen. They exhibit sexual dimorphism: females are about 2.5 millimeters (0.098 in) long; males are slightly smaller with darker backs. Males are easily distinguished from females based on colour differences, with a distinct black patch at the abdomen, less noticeable in recently emerged flies, and the sexcombs (a row of dark bristles on the tarsus of the first leg). Furthermore, males have a cluster of spiky hairs (claspers) surrounding the reproducing parts used to attach to the female during mating. There are extensive images at FlyBase.[29]
History of use in genetic analysis
D. melanogaster was among the first organisms used for genetic analysis, and today it is one of the most widely used and genetically best-known of all eukaryotic organisms. All organisms use common genetic systems; therefore, comprehending processes such as transcription and replication in fruit flies helps in understanding these processes in other eukaryotes, including humans.[30]
Thomas Hunt Morgan began using fruit flies in experimental studies of heredity at Columbia University in 1910 in a laboratory known as the Fly Room. The Fly Room was cramped with eight desks, each occupied by students and their experiments. They started off experiments using milk bottles to rear the fruit flies and handheld lenses for observing their traits. The lenses were later replaced by microscopes, which enhanced their observations. Morgan and his students eventually elucidated many basic principles of heredity, including sex-linked inheritance, epistasis, multiple alleles, and gene mapping.[30]
Model organism in genetics

D. melanogaster is one of the most studied organisms in biological research, particularly in genetics and developmental biology. The several reasons include:
- Its care and culture require little equipment, space, and expense even when using large cultures.
- It can be safely and readily anesthetized (usually with ether, carbon dioxide gas, by cooling, or with products like FlyNap).
- Its morphology is easy to identify once anesthetized.
- It has a short generation time (about 10 days at room temperature), so several generations can be studied within a few weeks.
- It has a high fecundity (females lay up to 100 eggs per day, and perhaps 2000 in a lifetime).[4]
- Males and females are readily distinguished, and virgin females are easily isolated, facilitating genetic crossing.
- The mature larvae has giant chromosomes in the salivary glands called polytene chromosomes—"puffs" indicate regions of transcription and hence gene activity.
- It has only four pairs of chromosomes: three autosomes, and one pair of sex chromosomes.
- Males do not show meiotic recombination, facilitating genetic studies.
- Recessive lethal "balancer chromosomes" carrying visible genetic markers can be used to keep stocks of lethal alleles in a heterozygous state without recombination due to multiple inversions in the balancer.
- The development of this organism—from fertilized egg to mature adult—is well understood.
- Genetic transformation techniques have been available since 1987.
- Its complete genome was sequenced and first published in 2000.[31]
- Sexual mosaics can be readily produced, providing an additional tool for studying the development and behavior of these flies.[32]
Genetic markers
Genetic markers are commonly used in Drosophila research, for example within balancer chromosomes or P-element inserts, and most phenotypes are easily identifiable either with the naked eye or under a microscope. In the list of a few common markers below, the allele symbol is followed by the name of the gene affected and a description of its phenotype. (Note: Recessive alleles are in lower case, while dominant alleles are capitalised.)
- Cy1: Curly; the wings curve away from the body, flight may be somewhat impaired.
- e1: ebony; black body and wings (heterozygotes are also visibly darker than wild type).
- Sb1: stubble; bristles are shorter and thicker than wild type.
- w1: white; eyes lack pigmentation and appear white.
- y1: yellow; body pigmentation and wings appear yellow. This is the fly analog of albinism.
Drosophila genes are traditionally named after the phenotype they cause when mutated. For example, the absence of a particular gene in Drosophila will result in a mutant embryo that does not develop a heart. Scientists have thus called this gene tinman, named after the Oz character of the same name.[33] Likewise changes in the Shavenbaby gene cause the loss of dorsal cuticular hairs in Drosophila sechellia larvae.[34] This system of nomenclature results in a wider range of gene names than in other organisms.
Genome
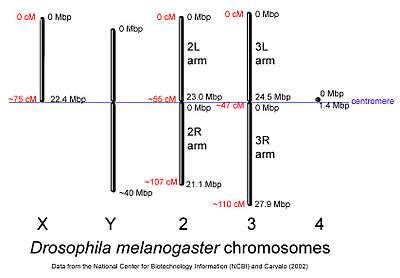
The genome of D. melanogaster (sequenced in 2000, and curated at the FlyBase database[31]) contains four pairs of chromosomes: an X/Y pair, and three autosomes labeled 2, 3, and 4. The fourth chromosome is so tiny, it is often ignored, aside from its important eyeless gene. The D. melanogaster sequenced genome of 139.5 million base pairs has been annotated[35] and contains around 15,682 genes according to Ensemble release 73. More than 60% of the genome appears to be functional non-protein-coding DNA[36] involved in gene expression control. Determination of sex in Drosophila occurs by the X:A ratio of X chromosomes to autosomes, not because of the presence of a Y chromosome as in human sex determination. Although the Y chromosome is entirely heterochromatic, it contains at least 16 genes, many of which are thought to have male-related functions.[37]
Similarity to humans
A March 2000 study by National Human Genome Research Institute comparing the fruit fly and human genome estimated that about 60% of genes are conserved between the two species.[38] About 75% of known human disease genes have a recognizable match in the genome of fruit flies,[39] and 50% of fly protein sequences have mammalian homologs. An online database called Homophila is available to search for human disease gene homologues in flies and vice versa.[40] Drosophila is being used as a genetic model for several human diseases including the neurodegenerative disorders Parkinson's, Huntington's, spinocerebellar ataxia and Alzheimer's disease.[41] The fly is also being used to study mechanisms underlying aging and oxidative stress, immunity, diabetes, and cancer, as well as drug abuse.[42][43][44]
Development
The life cycle of this insect has four stages: fertilized egg, larva, pupa, and adult.[5]
Embryogenesis in Drosophila has been extensively studied, as its small size, short generation time, and large brood size makes it ideal for genetic studies. It is also unique among model organisms in that cleavage occurs in a syncytium.

During oogenesis, cytoplasmic bridges called "ring canals" connect the forming oocyte to nurse cells. Nutrients and developmental control molecules move from the nurse cells into the oocyte. In the figure to the left, the forming oocyte can be seen to be covered by follicular support cells.
After fertilization of the oocyte, the early embryo (or syncytial embryo) undergoes rapid DNA replication and 13 nuclear divisions until about 5000 to 6000 nuclei accumulate in the unseparated cytoplasm of the embryo. By the end of the eighth division, most nuclei have migrated to the surface, surrounding the yolk sac (leaving behind only a few nuclei, which will become the yolk nuclei). After the 10th division, the pole cells form at the posterior end of the embryo, segregating the germ line from the syncytium. Finally, after the 13th division, cell membranes slowly invaginate, dividing the syncytium into individual somatic cells. Once this process is completed, gastrulation starts.[45]
Nuclear division in the early Drosophila embryo happens so quickly, no proper checkpoints exist, so mistakes may be made in division of the DNA. To get around this problem, the nuclei that have made a mistake detach from their centrosomes and fall into the centre of the embryo (yolk sac), which will not form part of the fly.
The gene network (transcriptional and protein interactions) governing the early development of the fruit fly embryo is one of the best understood gene networks to date, especially the patterning along the anteroposterior (AP) and dorsoventral (DV) axes (See under morphogenesis).[45]
The embryo undergoes well-characterized morphogenetic movements during gastrulation and early development, including germ-band extension, formation of several furrows, ventral invagination of the mesoderm, and posterior and anterior invagination of endoderm (gut), as well as extensive body segmentation until finally hatching from the surrounding cuticle into a first-instar larva.
During larval development, tissues known as imaginal discs grow inside the larva. Imaginal discs develop to form most structures of the adult body, such as the head, legs, wings, thorax, and genitalia. Cells of the imaginal disks are set aside during embryogenesis and continue to grow and divide during the larval stages—unlike most other cells of the larva, which have differentiated to perform specialized functions and grow without further cell division. At metamorphosis, the larva forms a pupa, inside which the larval tissues are reabsorbed and the imaginal tissues undergo extensive morphogenetic movements to form adult structures.
Sex determination
Drosophila flies have both X and Y chromosomes, as well as autosomes. Unlike humans, the Y chromosome does not confer maleness; rather, it encodes genes necessary for making sperm. Sex is instead determined by the ratio of X chromosomes to autosomes. Furthermore, each cell "decides" whether to be male or female independently of the rest of the organism, resulting in the occasional occurrence of gynandromorphs.
| X Chromosomes | Autosomes | Ratio of X:A | Sex |
|---|---|---|---|
| XXXX | AAAA | 1 | Normal Female |
| XXX | AAA | 1 | Normal Female |
| XXY | AA | 1 | Normal Female |
| XXYY | AA | 1 | Normal Female |
| XX | AA | 1 | Normal Female |
| XY | AA | 0.50 | Normal Male |
| X | AA | 0.50 | Normal Male (sterile) |
| XXX | AA | 1.50 | Metafemale |
| XXXX | AAA | 1.33 | Metafemale |
| XX | AAA | 0.66 | Intersex |
| X | AAA | 0.33 | Metamale |
Three major genes are involved in determination of Drosophila sex. These are sex-lethal, sisterless, and deadpan. Deadpan is an autosomal gene which inhibits sex-lethal, while sisterless is carried on the X chromosome and inhibits the action of deadpan. An AAX cell has twice as much deadpan as sisterless, so sex-lethal will be inhibited, creating a male. However, an AAXX cell will produce enough sisterless to inhibit the action of deadpan, allowing the sex-lethal gene to be transcribed to create a female.
Later, control by deadpan and sisterless disappears and what becomes important is the form of the sex-lethal gene. A secondary promoter causes transcription in both males and females. Analysis of the cDNA has shown that different forms are expressed in males and females. Sex-lethal has been shown to affect the splicing of its own mRNA. In males, the third exon is included which encodes a stop codon, causing a truncated form to be produced. In the female version, the presence of sex-lethal causes this exon to be missed out; the other seven amino acids are produced as a full peptide chain, again giving a difference between males and females.[46]
Presence or absence of functional sex-lethal proteins now go on to affect the transcription of another protein known as doublesex. In the absence of sex-lethal, doublesex will have the fourth exon removed and be translated up to and including exon 6 (DSX-M[ale]), while in its presence the fourth exon which encodes a stop codon will produce a truncated version of the protein (DSX-F[emale]). DSX-F causes transcription of Yolk proteins 1 and 2 in somatic cells, which will be pumped into the oocyte on its production.
Immunity
Unlike mammals, Drosophila flies only have innate immunity and lack an adaptive immune response. The D. melanogaster immune system can be divided into two responses: humoral and cell-mediated. The former is a systemic response mediated through the Toll and imd pathways, which are parallel systems for detecting microbes. The Toll pathway in Drosophila is known as the homologue of Toll-like pathways in mammals. Spatzle, a known ligand for the Toll pathway in flies, is produced in response to Gram-positive bacteria, parasites, and fungal infection. Upon infection, pro-Spatzle will be cleaved by protease SPE (Spatzle processing enzyme) to become active Spatzle, which then binds to the Toll receptor located on the cell surface (Fat body, hemocytes) and dimerise for activation of downstream NF-κB signaling pathways. The imd pathway, though, is triggered by Gram-negative bacteria through soluble and surface receptors (PGRP-LE and LC, respectively). D. melanogaster has a "fat body", which is thought to be homologous to the human liver. It is the primary secretory organ and produces antimicrobial peptides. These peptides are secreted into the hemolymph and bind infectious bacteria, killing them by forming pores in their cell walls. Other than the fat body, hemocytes, the blood cells in Drosophila, are known as the homologue of mammalian monocyte/macrophages, possessing a significant role in immune responses. It is known from the literature that in response to immune challenge, hemocytes are able to secrete cytokines, for example Spatzle, to activate downstream signaling pathways in the fat body. However, the mechanism still remains unclear. The response to infection can involve up to 2,423 genes, or 13.7% of the genome. Although the fly's transcriptional response to microbial challenge is highly specific to individual pathogens, Drosophila differentially expresses a core group of 252 genes upon infection with most bacteria. This core group of genes is associated with gene ontology categories such as antimicrobial response, stress response, secretion, neuron-like, reproduction, and metabolism among others.[47]
Behavioral genetics and neuroscience
In 1971, Ron Konopka and Seymour Benzer published "Clock mutants of Drosophila melanogaster", a paper describing the first mutations that affected an animal's behavior. Wild-type flies show an activity rhythm with a frequency of about a day (24 hours). They found mutants with faster and slower rhythms, as well as broken rhythms—flies that move and rest in random spurts. Work over the following 30 years has shown that these mutations (and others like them) affect a group of genes and their products that form a biochemical or biological clock. This clock is found in a wide range of fly cells, but the clock-bearing cells that control activity are several dozen neurons in the fly's central brain.
Since then, Benzer and others have used behavioral screens to isolate genes involved in vision, olfaction, audition, learning/memory, courtship, pain, and other processes, such as longevity.
Following the pioneering work of Alfred Henry Sturtevant [48] and others, Benzer and colleagues[32] used sexual mosaics to develop a novel fate mapping technique. This technique made it possible to assign a particular characteristic to a specific anatomical location. For example, this technique showed that male courtship behavior is controlled by the brain.[32] Mosaic fate mapping also provided the first indication of the existence of pheromones in this species.[49] Males distinguish between conspecific males and females and direct persistent courtship preferentially toward females thanks to a female-specific sex pheromone which is mostly produced by the female's tergites.
The first learning and memory mutants (dunce, rutabaga, etc.) were isolated by William "Chip" Quinn while in Benzer's lab, and were eventually shown to encode components of an intracellular signaling pathway involving cyclic AMP, protein kinase A, and a transcription factor known as CREB. These molecules were shown to be also involved in synaptic plasticity in Aplysia and mammals.[50]
Male flies sing to the females during courtship using their wings to generate sound, and some of the genetics of sexual behavior have been characterized. In particular, the fruitless gene has several different splice forms, and male flies expressing female splice forms have female-like behavior and vice versa. The TRP channels nompC, nanchung, and inactive are expressed in sound-sensitive Johnston's organ neurons and participate in the transduction of sound.[51][52]
The Nobel Prize in Physiology or Medicine for 2017 was awarded to Jeffrey C. Hall, Michael Rosbash, Michael W. Young for their works using fruit flies in understanding the "molecular mechanisms controlling the circadian rhythm".[53]
Transgenesis
It is now relatively simple to generate transgenic flies in Drosophila, relying on a variety of techniques. One approach of inserting foreign genes into the Drosophila genome involves P elements. The transposable P elements, also known as transposons, are segments of bacterial DNA that are transferred into the fly genome. Transgenic flies have already contributed to many scientific advances, e.g., modeling such human diseases as Parkinson's, neoplasia, obesity, and diabetes.[54]
Vision
The compound eye of the fruit fly contains 760 unit eyes or ommatidia, and are one of the most advanced among insects. Each ommatidium contains eight photoreceptor cells (R1-8), support cells, pigment cells, and a cornea. Wild-type flies have reddish pigment cells, which serve to absorb excess blue light so the fly is not blinded by ambient light.
Each photoreceptor cell consists of two main sections, the cell body and the rhabdomere. The cell body contains the nucleus, while the 100-μm-long rhabdomere is made up of toothbrush-like stacks of membrane called microvilli. Each microvillus is 1–2 μm in length and about 60 nm in diameter.[55] The membrane of the rhabdomere is packed with about 100 million rhodopsin molecules, the visual protein that absorbs light. The rest of the visual proteins are also tightly packed into the microvillar space, leaving little room for cytoplasm.
The photoreceptors in Drosophila express a variety of rhodopsin isoforms. The R1-R6 photoreceptor cells express rhodopsin1 (Rh1), which absorbs blue light (480 nm). The R7 and R8 cells express a combination of either Rh3 or Rh4, which absorb UV light (345 nm and 375 nm), and Rh5 or Rh6, which absorb blue (437 nm) and green (508 nm) light, respectively. Each rhodopsin molecule consists of an opsin protein covalently linked to a carotenoid chromophore, 11-cis-3-hydroxyretinal.[56]
As in vertebrate vision, visual transduction in invertebrates occurs via a G protein-coupled pathway. However, in vertebrates, the G protein is transducin, while the G protein in invertebrates is Gq (dgq in Drosophila). When rhodopsin (Rh) absorbs a photon of light its chromophore, 11-cis-3-hydroxyretinal, is isomerized to all-trans-3-hydroxyretinal. Rh undergoes a conformational change into its active form, metarhodopsin. Metarhodopsin activates Gq, which in turn activates a phospholipase Cβ (PLCβ) known as NorpA.[57]
PLCβ hydrolyzes phosphatidylinositol (4,5)-bisphosphate (PIP2), a phospholipid found in the cell membrane, into soluble inositol triphosphate (IP3) and diacylglycerol (DAG), which stays in the cell membrane. DAG or a derivative of DAG causes a calcium-selective ion channel known as transient receptor potential (TRP) to open and calcium and sodium flows into the cell. IP3 is thought to bind to IP3 receptors in the subrhabdomeric cisternae, an extension of the endoplasmic reticulum, and cause release of calcium, but this process does not seem to be essential for normal vision.[57]
Calcium binds to proteins such as calmodulin (CaM) and an eye-specific protein kinase C (PKC) known as InaC. These proteins interact with other proteins and have been shown to be necessary for shut off of the light response. In addition, proteins called arrestins bind metarhodopsin and prevent it from activating more Gq. A sodium-calcium exchanger known as CalX pumps the calcium out of the cell. It uses the inward sodium gradient to export calcium at a stoichiometry of 3 Na+/ 1 Ca++.[58]
TRP, InaC, and PLC form a signaling complex by binding a scaffolding protein called InaD. InaD contains five binding domains called PDZ domain proteins, which specifically bind the C termini of target proteins. Disruption of the complex by mutations in either the PDZ domains or the target proteins reduces the efficiency of signaling. For example, disruption of the interaction between InaC, the protein kinase C, and InaD results in a delay in inactivation of the light response.
Unlike vertebrate metarhodopsin, invertebrate metarhodopsin can be converted back into rhodopsin by absorbing a photon of orange light (580 nm).
About two-thirds of the Drosophila brain is dedicated to visual processing.[59] Although the spatial resolution of their vision is significantly worse than that of humans, their temporal resolution is around 10 times better.
Flight
The wings of a fly are capable of beating up to 220 times per second. Flies fly via straight sequences of movement interspersed by rapid turns called saccades.[60] During these turns, a fly is able to rotate 90° in less than 50 milliseconds.[60]
Characteristics of Drosophila flight may be dominated by the viscosity of the air, rather than the inertia of the fly body, but the opposite case with inertia as the dominant force may occur.[60] However, subsequent work showed that while the viscous effects on the insect body during flight may be negligible, the aerodynamic forces on the wings themselves actually cause fruit flies' turns to be damped viscously.[61]
As a pest
Drosophila is commonly considered a pest due to its tendency to infest habitations and establishments, where fermenting fruit is found; the flies may collect in homes, restaurants, stores, and other locations.[6] Removal of an infestation can be difficult, as the larvae may continue to hatch in nearby fermenting fruit even if the adult population is eliminated.
Misconception
The name and behaviour of this species of fly has led to the misconception that it is a biological security risk in Australia. While other "fruit fly" species do pose a risk, the D. melanogaster is attracted to fruit that is already rotting, rather than causing fruit to rot.[62][63]
See also
- Animal testing on invertebrates
- Eating behavior in Insects#Measurement
- Genetically modified insect
- Gynandromorphism
- Transgenesis
- Zebrafish - another widely used model organism in scientific research
References
- ↑ Meigen JW (1830). Systematische Beschreibung der bekannten europäischen zweiflügeligen Insekten. (Volume 6) (PDF) (in German). Schulz-Wundermann. Archived from the original (PDF) on 2010-02-01.
- ↑ "Vinegar Flies". Retrieved 19 July 2018.
- ↑ "Nobel Prizes".
- 1 2 Sang JH (2001-06-23). "Drosophila melanogaster: The Fruit Fly". In Reeve EC. Encyclopedia of genetics. USA: Fitzroy Dearborn Publishers, I. p. 157. ISBN 978-1-884964-34-3. Retrieved 2009-07-01.
- 1 2 Markow TA (June 2015). "The secret lives of Drosophila flies". eLife. 4. doi:10.7554/eLife.06793. PMC 4454838. PMID 26041333.
- 1 2 "Vinegar Flies, Drosophila species, Family: Drosophilidae". Department of Entomology, College of Agricultural Sciences, Pennsylvania State University. 2017. Retrieved 20 July 2017.
- ↑ McGurk, L; Berson, A; Bonini, N. M (2015). "Drosophila as an in Vivo Model for Human Neurodegenerative Disease". Genetics. 201 (2): 377–402. doi:10.1534/genetics.115.179457. PMC 4596656. PMID 26447127.
- 1 2 3 4 5 6 7 Ashburner M, Thompson JN (1978). "The laboratory culture of Drosophila". In Ashburner M, Wright TRF. The genetics and biology of Drosophila. 2A. Academic Press. 1–81.
- 1 2 3 4 5 6 7 Ashburner M, Golic KG, Hawley RS (2005). Drosophila: A Laboratory Handbook (2nd ed.). Cold Spring Harbor Laboratory Press. pp. 162–4. ISBN 978-0-87969-706-8.
- ↑ Bloomington Drosophila Stock Center at Indiana University: Basic Methods of Culturing Drosophila
- 1 2 Chiang HC, Hodson AC (1950). "An analytical study of population growth in Drosophila melanogaster". Ecological Monographs. 20 (3): 173–206. doi:10.2307/1948580. JSTOR 1948580.
- ↑ Bakker K (1961). "An analysis of factors which determine success in competition for food among larvae of Drosophila melanogaster". Archives Neerlandaises de Zoologie. 14 (2): 200–281. doi:10.1163/036551661X00061.
- ↑ Blum JE, Fischer CN, Miles J, Handelsman J (November 2013). "Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster". MBio. 4 (6): e00860–13. doi:10.1128/mBio.00860-13. PMC 3892787. PMID 24194543.
- ↑ Koerth-Baker M (August 21, 2009). "Female Flies Put Up a Fight to Keep Sex Short". National Geographic News. Retrieved August 21, 2009.
- ↑ Connolly K, Cook R (1973). "Rejection Responses by Female Drosophila melanogaster: Their Ontogeny, Causality and Effects upon the Behaviour of the Courting Male". Behaviour. 44 (1/2): 142–166. doi:10.1163/156853973x00364. JSTOR 4533484.
- ↑ Houot B, Svetec N, Godoy-Herrera R, Ferveur JF (July 2010). "Effect of laboratory acclimation on the variation of reproduction-related characters in Drosophila melanogaster". The Journal of Experimental Biology. 213 (Pt 13): 2322–31. doi:10.1242/jeb.041566. PMID 20543131.
- ↑ Gilbert SF (2006). "9: Fertilization in Drosophila". In 8th. Developmental Biology. Sinauer Associates. ISBN 978-0-87893-250-4. Archived from the original on 2007-02-07.
- 1 2 3 Price CS, Dyer KA, Coyne JA (July 1999). "Sperm competition between Drosophila males involves both displacement and incapacitation". Nature. 400 (6743): 449–52. Bibcode:1999Natur.400..449P. doi:10.1038/22755. PMID 10440373.
- 1 2 3 4 "Fruit fly research may reveal what happens in female brains during courtship, mating". Retrieved October 5, 2014.
- ↑ Carnes MU, Campbell T, Huang W, Butler DG, Carbone MA, Duncan LH, Harbajan SV, King EM, Peterson KR, Weitzel A, Zhou S, Mackay TF (2015). "The Genomic Basis of Postponed Senescence in Drosophila melanogaster". PLOS One. 10 (9): e0138569. Bibcode:2015PLoSO..1038569C. doi:10.1371/journal.pone.0138569. PMC 4574564. PMID 26378456.
- ↑ Loyau A, Cornuau JH, Clobert J, Danchin E (2012). "Incestuous sisters: mate preference for brothers over unrelated males in Drosophila melanogaster". PLOS One. 7 (12): e51293. Bibcode:2012PLoSO...751293L. doi:10.1371/journal.pone.0051293. PMC 3519633. PMID 23251487.
- ↑ Pitnick S (1996). "Investment in testes and the cost of making long sperm in Drosophila". American Naturalist. 148: 57–80. doi:10.1086/285911.
- ↑ Dagaeff AC, Pocheville A, Nöbel S, Loyau A, Isabel G, Danchin E (2016). "Drosophila mate copying correlates with atmospheric pressure in a speed learning situation". Animal Behaviour. 121: 163–174. doi:10.1016/j.anbehav.2016.08.022.
- ↑ Dukas, Reuven (2004). "Male fruit flies learn to avoid interspecific courtship". Behavioral Ecology. 15 (4): 695–698. doi:10.1093/beheco/arh068.
- ↑ Saleem S, Ruggles PH, Abbott WK, Carney GE (2014). "Sexual experience enhances Drosophila melanogaster male mating behavior and success". PLOS One. 9 (5): e96639. Bibcode:2014PLoSO...996639S. doi:10.1371/journal.pone.0096639. PMC 4013029. PMID 24805129.
- 1 2 Haartman, Lars von (1951). "Successive Polygamy". Behaviour. 3 (1): 256–273. doi:10.1163/156853951x00296.
- 1 2 3 4 5 6 7 Vartak VR, Varma V, Sharma VK (February 2015). "Effects of polygamy on the activity/rest rhythm of male fruit flies Drosophila melanogaster". Die Naturwissenschaften. 102 (1–2): 1252. Bibcode:2015SciNa.102....3V. doi:10.1007/s00114-014-1252-5. PMID 25604736.
- 1 2 Bateman AJ (December 1948). "Intra-sexual selection in Drosophila". Heredity. 2 (Pt. 3): 349–68. doi:10.1038/hdy.1948.21. PMID 18103134.
- ↑ "FlyBase: A database of Drosophila genes and genomes". Genetics Society of America. 2009. Archived from the original on August 15, 2009. Retrieved August 11, 2009.
- 1 2 Pierce BA (2004). Genetics: A Conceptual Approach (2nd ed.). W. H. Freeman. ISBN 978-0-7167-8881-2.
- 1 2 Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. (March 2000). "The genome sequence of Drosophila melanogaster". Science. 287 (5461): 2185–95. Bibcode:2000Sci...287.2185.. doi:10.1126/science.287.5461.2185. PMID 10731132.
- 1 2 3 Hotta Y, Benzer S (December 1972). "Mapping of behaviour in Drosophila mosaics". Nature. 240 (5383): 527–35. Bibcode:1972Natur.240..527H. doi:10.1038/240527a0. PMID 4568399.
- ↑ Azpiazu N, Frasch M (July 1993). "tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila". Genes & Development. 7 (7B): 1325–40. doi:10.1101/gad.7.7b.1325. PMID 8101173.
- ↑ Stern, D. L.; Frankel, N. (11 November 2013). "The structure and evolution of cis-regulatory regions: the shavenbaby story". Philosophical Transactions of the Royal Society B: Biological Sciences. 368 (1632): 20130028–20130028. doi:10.1098/rstb.2013.0028. PMC 3826501. PMID 24218640.
- ↑ "NCBI (National Center for Biotechnology Information) Genome Database". Retrieved 2011-11-30.
- ↑ Halligan DL, Keightley PD (July 2006). "Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison". Genome Research. 16 (7): 875–84. doi:10.1101/gr.5022906. PMC 1484454. PMID 16751341.
- ↑ Carvalho AB (December 2002). "Origin and evolution of the Drosophila Y chromosome". Current Opinion in Genetics & Development. 12 (6): 664–8. doi:10.1016/S0959-437X(02)00356-8. PMID 12433579.
- ↑ "Background on Comparative Genomic Analysis". US National Human Genome Research Institute. December 2002.
- ↑ Reiter LT, Potocki L, Chien S, Gribskov M, Bier E (June 2001). "A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster". Genome Research. 11 (6): 1114–25. doi:10.1101/gr.169101. PMC 311089. PMID 11381037.
- ↑ Chien S, Reiter LT, Bier E, Gribskov M (January 2002). "Homophila: human disease gene cognates in Drosophila". Nucleic Acids Research. National Library of Medicine (NLM). 30 (1): 149–51. doi:10.1093/nar/30.1.149. PMC 99119. PMID 11752278.
- ↑ Jaiswal M, Sandoval H, Zhang K, Bayat V, Bellen HJ (2012). "Probing mechanisms that underlie human neurodegenerative diseases in Drosophila". Annual Review of Genetics. 46: 371–96. doi:10.1146/annurev-genet-110711-155456. PMC 3663445. PMID 22974305.
- ↑ Pick L (2017). Fly Models of Human Diseases. Volume 121 of Current Topics in Developmental Biology. Academic Press. ISBN 978-0-12-802905-3.
- ↑ Buchon N, Silverman N, Cherry S (December 2014). "Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology". Nature Reviews. Immunology. 14 (12): 796–810. doi:10.1038/nri3763. PMID 25421701.
- ↑ Kaun KR, Devineni AV, Heberlein U (June 2012). "Drosophila melanogaster as a model to study drug addiction". Human Genetics. 131 (6): 959–75. doi:10.1007/s00439-012-1146-6. PMC 3351628. PMID 22350798.
- 1 2 Weigmann K, Klapper R, Strasser T, Rickert C, Technau G, Jäckle H, Janning W, Klämbt C (June 2003). "FlyMove--a new way to look at development of Drosophila". Trends in Genetics. 19 (6): 310–1. doi:10.1016/S0168-9525(03)00050-7. PMID 12801722.
- ↑ Gilbert SF (2000). Developmental Biology (6th ed.). Sunderland (MA): Sinauer Associates; 2000.
- ↑ Troha K, Im JH, Revah J, Lazzaro BP, Buchon N (February 2018). "Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster". PLoS Pathogens. 14 (2): e1006847. doi:10.1371/journal.ppat.1006847. PMC 5812652. PMID 29394281.
- ↑ Sturtevant AH (1929). "The claret mutant type of Drosophila simulans: a study of chromosome elimination and cell-lineage". Zeitschrift für wissenschaftliche Zoologie. 135: 323–356.
- ↑ Nissani M (May 1975). "A new behavioral bioassay for an analysis of sexual attraction and pheromones in insects". The Journal of Experimental Zoology. 192 (2): 271–5. doi:10.1002/jez.1401920217. PMID 805823.
- ↑ Khan FA (2011). Biotechnology Fundamentals. CRC Press. p. 213. ISBN 978-1-4398-2009-4.
- ↑ Lehnert BP, Baker AE, Gaudry Q, Chiang AS, Wilson RI (January 2013). "Distinct roles of TRP channels in auditory transduction and amplification in Drosophila". Neuron. 77 (1): 115–28. doi:10.1016/j.neuron.2012.11.030. PMC 3811118. PMID 23312520.
- ↑ Zhang W, Yan Z, Jan LY, Jan YN (August 2013). "Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae". Proceedings of the National Academy of Sciences of the United States of America. 110 (33): 13612–7. Bibcode:2013PNAS..11013612Z. doi:10.1073/pnas.1312477110. PMC 3746866. PMID 23898199.
- ↑ "The 2017 Nobel Prize in Physiology or Medicine jointly to Jeffrey C. Hall, Michael Rosbash and Michael W. Young for their discoveries of molecular mechanisms controlling the circadian rhythm". Nobelprize.org. 2 October 2017. Retrieved 5 October 2017.
- ↑ Hughes TT, Allen AL, Bardin JE, Christian MN, Daimon K, Dozier KD, Hansen CL, Holcomb LM, Ahlander J (February 2012). "Drosophila as a genetic model for studying pathogenic human viruses". Virology. 423 (1): 1–5. doi:10.1016/j.virol.2011.11.016. PMC 3253880. PMID 22177780.
- ↑ Hardie RC, Raghu P (September 2001). "Visual transduction in Drosophila". Nature. 413 (6852): 186–93. Bibcode:2001Natur.413..186H. doi:10.1038/35093002. PMID 11557987.
- ↑ Nichols R, Pak WL (October 1985). "Characterization of Drosophila melanogaster rhodopsin". The Journal of Biological Chemistry. 260 (23): 12670–4. PMID 3930500.
- 1 2 Raghu P, Colley NJ, Webel R, James T, Hasan G, Danin M, Selinger Z, Hardie RC (May 2000). "Normal phototransduction in Drosophila photoreceptors lacking an InsP(3) receptor gene". Molecular and Cellular Neurosciences. 15 (5): 429–45. doi:10.1006/mcne.2000.0846. PMID 10833300.
- ↑ Wang T, Xu H, Oberwinkler J, Gu Y, Hardie RC, Montell C (February 2005). "Light activation, adaptation, and cell survival functions of the Na+/Ca2+ exchanger CalX". Neuron. 45 (3): 367–78. doi:10.1016/j.neuron.2004.12.046. PMID 15694324.
- ↑ Rein K, Zöckler M, Mader MT, Grübel C, Heisenberg M (February 2002). "The Drosophila standard brain". Current Biology. 12 (3): 227–31. doi:10.1016/S0960-9822(02)00656-5. PMID 11839276.
- 1 2 3 Fry SN, Sayaman R, Dickinson MH (April 2003). "The aerodynamics of free-flight maneuvers in Drosophila" (PDF). Science. 300 (5618): 495–8. Bibcode:2003Sci...300..495F. doi:10.1126/science.1081944. PMID 12702878. Archived from the original (PDF) on 2015-09-24.
- ↑ Hesselberg T, Lehmann FO (December 2007). "Turning behaviour depends on frictional damping in the fruit fly Drosophila". The Journal of Experimental Biology. 210 (Pt 24): 4319–34. doi:10.1242/jeb.010389. PMID 18055621.
- ↑ "Non pest species". Plant Health Australia. Retrieved September 19, 2017.
- ↑ McEvey S (February 5, 2014). "Fruit Flies: A Case Of Mistaken Identity". Australian Museum. Retrieved September 19, 2017.
Further reading
- Kohler RE (1994). Lords of the Fly: Drosophila genetics and the experimental life. Chicago: University of Chicago Press. ISBN 978-0-226-45063-6.
- Gilbert SF (2000). Developmental Biology (6th ed.). Sunderland (MA): Sinauer Associates; 2000.
- Perrimon N, Bonini NM, Dhillon P (March 2016). "Fruit flies on the front line: the translational impact of Drosophila". Disease Models & Mechanisms. 9 (3): 229–31. doi:10.1242/dmm.024810. PMC 4833334. PMID 26935101.
- Henderson M (April 8, 2010). "Row over fruit fly Drosophila melanogaster name bugs scientists". The Times. The Australian. Retrieved September 19, 2017.
Popular media
- Part 1 of the "Small fly: BIG impact" educational videos explaining the history and importance of the model organism Drosophila.
- Part 2 of the "Small fly: BIG impact" educational videos explaining how research is carried out in Drosophila.
- "Inside the Fly Lab" — broadcast by WGBH and PBS, in the program series "Curious", January 2008.
- "How a Fly Detects Poison" — WhyFiles.org article describes how the fruit fly tastes a larva-killing chemical in food.
External links
| Wikispecies has information related to Drosophila melanogaster |
| Wikimedia Commons has media related to Drosophila melanogaster. |
- "A quick and simple introduction to Drosophila melanogaster". Drosophila Virtual Library.
- "FlyBase — A Database of Drosophila Genes & Genomes".
- "NCBI Map Viewer - Drosophila melanogaster".
- "Drosophila Virtual Library".
- "The Berkeley Drosophila Genome Project".
- "FlyMove". - video resources for Drosophila development
- "Drosophila Nomenclature — naming of genes". Archived from the original on 8 October 2011.
- View the Fruitfly genome on Ensembl
- View the dm6 genome assembly in the UCSC Genome Browser.
- Manchester Fly Facility - for the public from the University of Manchester
- The droso4schools website with school-relevant resources about Drosophila: