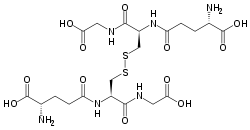
Glutathione disulfide
 | |
| Names | |
|---|---|
| IUPAC name
(2S)-2-Amino-5-[[(2R)-3-[(2R)-2-[[(4S)-4-amino-5-hydroxy-5-oxopentanoyl]amino]-3-(carboxymethylamino)-3-oxopropyl]disulfanyl-1-
(carboxymethylamino)-1-oxopropan-2-yl]amino]-5-oxopentanoic acid | |
| Identifiers | |
3D model (JSmol) |
|
| Abbreviations | GSSG |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.043.777 |
PubChem CID |
|
| |
| |
| Properties | |
| C20H32N6O12S2 | |
| Molar mass | 612.63 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Glutathione disulfide (GSSG) is a disulfide derived from two glutathione molecules.[1]
In living cells, glutathione disulfide is reduced into two molecules of glutathione with reducing equivalents from the coenzyme NADPH. This reaction is catalyzed by the enzyme glutathione reductase.[2] Antioxidant enzymes, such as glutathione peroxidases and peroxiredoxins, generate glutathione disulfide during the reduction of peroxides such as hydrogen peroxide (H2O2) and organic hydroperoxides (ROOH):[3]
- 2 GSH + ROOH → GSSG + ROH + H2O
Other enzymes, such as glutaredoxins, generate glutathione disulfide through thiol-disulfide exchange with protein disulfide bonds or other low molecular mass compounds, such as coenzyme A disulfide or dehydroascorbic acid.[4]
- 2 GSH + R-S-S-R → GSSG + 2 RSH
The GSH:GSSG ratio is therefore an important bioindicator of cellular health, with a higher ratio signifying less oxidative stress in the organism. A lower ratio may even be indicative of neurodegenerative diseases, such as Parkinson's disease (PD) and Alzheimer's disease.[5]
Neuromodulator
GSSG, along with glutathione and S-nitrosoglutathione (GSNO), have been found to bind to the glutamate recognition site of the NMDA and AMPA receptors (via their γ-glutamyl moieties), and may be endogenous neuromodulators.[6][7] At millimolar concentrations, they may also modulate the redox state of the NMDA receptor complex.[7]
See also
References
- ↑ Meister A, Anderson ME (1983). "Glutathione". Annual Review of Biochemistry. 52: 711–60. doi:10.1146/annurev.bi.52.070183.003431. PMID 6137189.
- ↑ Deneke SM, Fanburg BL (1989). "Regulation of cellular glutathione". The American Journal of Physiology. 257 (4 Pt 1): L163–73. PMID 2572174.
- ↑ Meister A (1988). "Glutathione metabolism and its selective modification". The Journal of Biological Chemistry. 263 (33): 17205–8. PMID 3053703.
- ↑ Holmgren A, Johansson C, Berndt C, Lönn ME, Hudemann C, Lillig CH (December 2005). "Thiol redox control via thioredoxin and glutaredoxin systems". Biochem. Soc. Trans. 33 (Pt 6): 1375–7. doi:10.1042/BST20051375. PMID 16246122.
- ↑ Owen, Joshua B.; Butterfield, D. Allan (2010). "Measurement of oxidized/reduced glutathione ratio". In Bross, Peter; Gregersen, Niels. Protein Misfolding and Cellular Stress in Disease and Aging. Methods in Molecular Biology. 648. pp. 269–77. doi:10.1007/978-1-60761-756-3_18. ISBN 978-1-60761-755-6. PMID 20700719.
- ↑ Steullet P, Neijt HC, Cuénod M, Do KQ (2006). "Synaptic plasticity impairment and hypofunction of NMDA receptors induced by glutathione deficit: relevance to schizophrenia". Neuroscience. 137 (3): 807–19. doi:10.1016/j.neuroscience.2005.10.014. PMID 16330153.
- 1 2 Varga V, Jenei Z, Janáky R, Saransaari P, Oja SS (1997). "Glutathione is an endogenous ligand of rat brain N-methyl-D-aspartate (NMDA) and 2-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) receptors". Neurochemical Research. 22 (9): 1165–71. doi:10.1023/A:1027377605054. PMID 9251108.