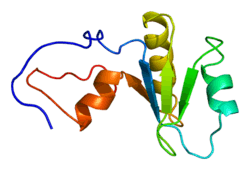Poly(A)-specific ribonuclease
Poly(A)-specific ribonuclease (PARN), also known as polyadenylate-specific ribonuclease or deadenylating nuclease (DAN), is an enzyme that in humans is encoded by the PARN gene.[5][6]
Function
Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs. The amino acid sequence of poly(A)-specific ribonuclease shows homology to the RNase D family of 3'-exonucleases. The protein appears to be localized in both the nucleus and the cytoplasm. It is not stably associated with polysomes or ribosomal subunits.[6] Hereditary mutations in PARN lead to the bone marrow failure disease Dyskeratosis Congenita which is caused by defective telomerase RNA processing and degradation in patients[7][8][9][10][11][12][13].
References
- 1 2 3 ENSG00000274829 GRCh38: Ensembl release 89: ENSG00000140694, ENSG00000274829 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000022685 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Buiting K, Körner C, Ulrich B, Wahle E, Horsthemke B (May 2000). "Prader-Willi". Cytogenetics and Cell Genetics. 87 (1–2): 125–31. doi:10.1159/000015378. PMID 10640832.
- 1 2 "Entrez Gene: PARN poly(A)-specific ribonuclease (deadenylation nuclease)".
- ↑ Tummala H, Walne A, Collopy L, Cardoso S, de la Fuente J, Lawson S, Powell J, Cooper N, Foster A, Mohammed S, Plagnol V, Vulliamy T, Dokal I (May 2015). "Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita". The Journal of Clinical Investigation. 125 (5): 2151–60. doi:10.1172/JCI78963. PMC 4463202. PMID 25893599.
- ↑ Dhanraj S, Gunja SM, Deveau AP, Nissbeck M, Boonyawat B, Coombs AJ, Renieri A, Mucciolo M, Marozza A, Buoni S, Turner L, Li H, Jarrar A, Sabanayagam M, Kirby M, Shago M, Pinto D, Berman JN, Scherer SW, Virtanen A, Dror Y (November 2015). "Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN)". Journal of Medical Genetics. 52 (11): 738–48. doi:10.1136/jmedgenet-2015-103292. PMID 26342108.
- ↑ Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, Weissler J, Fitzgerald J, Kershaw C, Klesney-Tait J, Mageto Y, Shay JW, Ji W, Bilguvar K, Mane S, Lifton RP, Garcia CK (May 2015). "Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening". Nature Genetics. 47 (5): 512–7. doi:10.1038/ng.3278. PMC 4414891. PMID 25848748.
- ↑ Shukla S, Schmidt JC, Goldfarb KC, Cech TR, Parker R (April 2016). "Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects". Nature Structural & Molecular Biology. 23 (4): 286–92. doi:10.1038/nsmb.3184. PMC 4830462. PMID 26950371.
- ↑ Tseng CK, Wang HF, Burns AM, Schroeder MR, Gaspari M, Baumann P (December 2015). "Human Telomerase RNA Processing and Quality Control". Cell Reports. 13 (10): 2232–43. doi:10.1016/j.celrep.2015.10.075. PMID 26628367.
- ↑ Nguyen D, Grenier St-Sauveur V, Bergeron D, Dupuis-Sandoval F, Scott MS, Bachand F (December 2015). "A Polyadenylation-Dependent 3' End Maturation Pathway Is Required for the Synthesis of the Human Telomerase RNA". Cell Reports. 13 (10): 2244–57. doi:10.1016/j.celrep.2015.11.003. PMID 26628368.
- ↑ Moon DH, Segal M, Boyraz B, Guinan E, Hofmann I, Cahan P, Tai AK, Agarwal S (December 2015). "Poly(A)-specific ribonuclease (PARN) mediates 3'-end maturation of the telomerase RNA component". Nature Genetics. 47 (12): 1482–8. doi:10.1038/ng.3423. PMC 4791094. PMID 26482878.
Further reading
- Körner CG, Wahle E (April 1997). "Poly(A) tail shortening by a mammalian poly(A)-specific 3'-exoribonuclease". The Journal of Biological Chemistry. 272 (16): 10448–56. doi:10.1074/jbc.272.16.10448. PMID 9099687.
- Körner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E (September 1998). "The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes". The EMBO Journal. 17 (18): 5427–37. doi:10.1093/emboj/17.18.5427. PMC 1170868. PMID 9736620.
- Dehlin E, Wormington M, Körner CG, Wahle E (March 2000). "Cap-dependent deadenylation of mRNA". The EMBO Journal. 19 (5): 1079–86. doi:10.1093/emboj/19.5.1079. PMC 305646. PMID 10698948.
- Martinez J, Ren YG, Thuresson AC, Hellman U, Astrom J, Virtanen A (August 2000). "A 54-kDa fragment of the Poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting Poly(A)-specific 3' exonuclease". The Journal of Biological Chemistry. 275 (31): 24222–30. doi:10.1074/jbc.M001705200. PMID 10801819.
- Gao M, Fritz DT, Ford LP, Wilusz J (March 2000). "Interaction between a poly(A)-specific ribonuclease and the 5' cap influences mRNA deadenylation rates in vitro". Molecular Cell. 5 (3): 479–88. doi:10.1016/S1097-2765(00)80442-6. PMC 2811581. PMID 10882133.
- Gao M, Wilusz CJ, Peltz SW, Wilusz J (March 2001). "A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements". The EMBO Journal. 20 (5): 1134–43. doi:10.1093/emboj/20.5.1134. PMC 145468. PMID 11230136.
- Martînez J, Ren YG, Nilsson P, Ehrenberg M, Virtanen A (July 2001). "The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation". The Journal of Biological Chemistry. 276 (30): 27923–9. doi:10.1074/jbc.M102270200. PMID 11359775.
- Copeland PR, Wormington M (June 2001). "The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN". Rna. 7 (6): 875–86. doi:10.1017/S1355838201010020. PMC 1370141. PMID 11424938.
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M (November 2001). "AU binding proteins recruit the exosome to degrade ARE-containing mRNAs". Cell. 107 (4): 451–64. doi:10.1016/S0092-8674(01)00578-5. PMID 11719186.
- Ren YG, Martínez J, Virtanen A (February 2002). "Identification of the active site of poly(A)-specific ribonuclease by site-directed mutagenesis and Fe(2+)-mediated cleavage". The Journal of Biological Chemistry. 277 (8): 5982–7. doi:10.1074/jbc.M111515200. PMID 11742007.
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI (January 2002). "Directed proteomic analysis of the human nucleolus". Current Biology. 12 (1): 1–11. doi:10.1016/S0960-9822(01)00650-9. PMID 11790298.
- Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ (November 2002). "Functional proteomic analysis of human nucleolus". Molecular Biology of the Cell. 13 (11): 4100–9. doi:10.1091/mbc.E02-05-0271. PMC 133617. PMID 12429849.
- Lai WS, Kennington EA, Blackshear PJ (June 2003). "Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease". Molecular and Cellular Biology. 23 (11): 3798–812. doi:10.1128/MCB.23.11.3798-3812.2003. PMC 155217. PMID 12748283.
- Lejeune F, Li X, Maquat LE (September 2003). "Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities". Molecular Cell. 12 (3): 675–87. doi:10.1016/S1097-2765(03)00349-6. PMID 14527413.
- Gherzi R, Lee KY, Briata P, Wegmüller D, Moroni C, Karin M, Chen CY (June 2004). "A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery". Molecular Cell. 14 (5): 571–83. doi:10.1016/j.molcel.2004.05.002. PMID 15175153.
- Lehner B, Sanderson CM (July 2004). "A protein interaction framework for human mRNA degradation". Genome Research. 14 (7): 1315–23. doi:10.1101/gr.2122004. PMC 442147. PMID 15231747.
- Ren YG, Kirsebom LA, Virtanen A (November 2004). "Coordination of divalent metal ions in the active site of poly(A)-specific ribonuclease". The Journal of Biological Chemistry. 279 (47): 48702–6. doi:10.1074/jbc.M403858200. PMID 15358788.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.