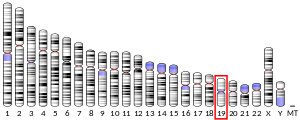
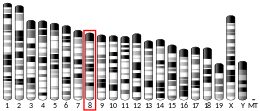
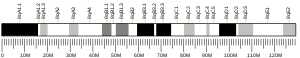
RNASEH2A
Ribonuclease H2 subunit A, also known as RNase H2 subunit A, is an enzyme that in humans is encoded by the RNASEH2A gene.[5]
Function
The protein encoded by this gene is a component of the heterotrimeric type II ribonuclease H enzyme (RNaseH2). The other two subunits are the non-catalytic RNASEH2B and RNASEH2C. RNaseH2 is the major source of ribonuclease H activity in mammalian cells and endonucleolytically cleaves ribonucleotides. It is predicted to remove Okazaki fragment RNA primers during lagging strand DNA synthesis and to excise single ribonucleotides from DNA-DNA duplexes.[5]
Clinical significance
Mutations in this gene cause Aicardi-Goutieres syndrome (AGS), an autosomal recessive neurological disorder characterized by progressive microcephaly and psychomotor retardation, intracranial calcifications, elevated levels of interferon-alpha and white blood cells in the cerebrospinal fluid.[5]
References
Further reading
- Crow YJ, Leitch A, Hayward BE, et al. (2006). "Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection". Nat. Genet. 38 (8): 910–6. doi:10.1038/ng1842. PMID 16845400.
- Chon H, Vassilev A, DePamphilis ML, et al. (2009). "Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex". Nucleic Acids Res. 37 (1): 96–110. doi:10.1093/nar/gkn913. PMC 2615623. PMID 19015152.
- Flanagan JM, Funes JM, Henderson S, et al. (2009). "Genomics screen in transformed stem cells reveals RNASEH2A, PPAP2C, and ADARB1 as putative anticancer drug targets". Mol. Cancer Ther. 8 (1): 249–60. doi:10.1158/1535-7163.MCT-08-0636. PMID 19139135.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Bonaldo MF, Lennon G, Soares MB (1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Res. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Rice G, Patrick T, Parmar R, et al. (2007). "Clinical and Molecular Phenotype of Aicardi-Goutières Syndrome". Am. J. Hum. Genet. 81 (4): 713–25. doi:10.1086/521373. PMC 2227922. PMID 17846997.
- Frank P, Braunshofer-Reiter C, Wintersberger U, et al. (1998). "Cloning of the cDNA encoding the large subunit of human RNase HI, a homologue of the prokaryotic RNase HII". Proc. Natl. Acad. Sci. U.S.A. 95 (22): 12872–7. doi:10.1073/pnas.95.22.12872. PMC 23637. PMID 9789007.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2002). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Ganesh SK, Zakai NA, van Rooij FJ, et al. (2009). "Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium". Nat. Genet. 41 (11): 1191–8. doi:10.1038/ng.466. PMC 2778265. PMID 19862010.
External links
- GeneReviews/NCBI/NIH/UW entry on Aicardi-Goutières Syndrome
- OMIM entries on Aicardi-Goutieres syndrome