RNASEH1
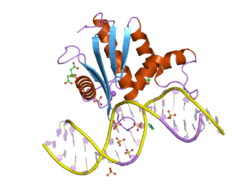
Ribonuclease H1 also known as RNase H1 is an enzyme that in humans is encoded by the RNASEH1 gene.[5][6][7] The RNase H1 is a non-specific endonuclease and catalyzes the cleavage of RNA via a hydrolytic mechanism.
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000171865 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000020630 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ "Entrez Gene: ribonuclease H1".
- ↑ ten Asbroek AL, van Groenigen M, Jakobs ME, Koevoets C, Janssen B, Baas F (June 2002). "Ribonuclease H1 maps to chromosome 2 and has at least three pseudogene loci in the human genome". Genomics. 79 (6): 818–23. doi:10.1006/geno.2002.6776. PMID 12036296.
- ↑ Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W (October 2007). "Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription". Mol. Cell. 28 (2): 264–76. doi:10.1016/j.molcel.2007.08.015. PMID 17964265.
Further reading
- Nowotny M, Cerritelli SM, Ghirlando R, et al. (2008). "Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD". EMBO J. 27 (7): 1172–81. doi:10.1038/emboj.2008.44. PMC 2323259. PMID 18337749.
- Lima WF, Wu H, Nichols JG, et al. (2003). "Human RNase H1 uses one tryptophan and two lysines to position the enzyme at the 3'-DNA/5'-RNA terminus of the heteroduplex substrate". J. Biol. Chem. 278 (50): 49860–7. doi:10.1074/jbc.M306543200. PMID 14506260.
- Lima WF, Rose JB, Nichols JG, et al. (2007). "The positional influence of the helical geometry of the heteroduplex substrate on human RNase H1 catalysis". Mol. Pharmacol. 71 (1): 73–82. doi:10.1124/mol.106.025429. PMID 17028157.
- Lima WF, Nichols JG, Wu H, et al. (2004). "Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis". J. Biol. Chem. 279 (35): 36317–26. doi:10.1074/jbc.M405035200. PMID 15205459.
- Otsuki T, Ota T, Nishikawa T, et al. (2005). "Signal sequence and keyword trap in silico for selection of full-length human cDNAs encoding secretion or membrane proteins from oligo-capped cDNA libraries". DNA Res. 12 (2): 117–26. doi:10.1093/dnares/12.2.117. PMID 16303743.
- Wu H, Lima WF, Crooke ST (1999). "Properties of cloned and expressed human RNase H1". J. Biol. Chem. 274 (40): 28270–8. doi:10.1074/jbc.274.40.28270. PMID 10497183.
- Frank P, Braunshofer-Reiter C, Pöltl A, Holzmann K (1998). "Cloning, subcellular localization and functional expression of human RNase HII". Biol. Chem. 379 (12): 1407–12. doi:10.1515/bchm.1998.379.12.1407. PMID 9894807.
- Wu H, Lima WF, Zhang H, et al. (2004). "Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs". J. Biol. Chem. 279 (17): 17181–9. doi:10.1074/jbc.M311683200. PMID 14960586.
- Hillier LW, Graves TA, Fulton RS, et al. (2005). "Generation and annotation of the DNA sequences of human chromosomes 2 and 4". Nature. 434 (7034): 724–31. doi:10.1038/nature03466. PMID 15815621.
- Lima WF, Wu H, Nichols JG, et al. (2003). "Human RNase H1 activity is regulated by a unique redox switch formed between adjacent cysteines". J. Biol. Chem. 278 (17): 14906–12. doi:10.1074/jbc.M211279200. PMID 12473655.
- ten Asbroek AL, van Groenigen M, Nooij M, Baas F (2002). "The involvement of human ribonucleases H1 and H2 in the variation of response of cells to antisense phosphorothioate oligonucleotides". Eur. J. Biochem. 269 (2): 583–92. doi:10.1046/j.0014-2956.2001.02686.x. PMID 11856317.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Cerritelli SM, Crouch RJ (1998). "Cloning, expression, and mapping of ribonucleases H of human and mouse related to bacterial RNase HI". Genomics. 53 (3): 300–7. doi:10.1006/geno.1998.5497. PMID 9799596.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2002). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Cerritelli SM, Frolova EG, Feng C, et al. (2003). "Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice". Mol. Cell. 11 (3): 807–15. doi:10.1016/S1097-2765(03)00088-1. PMID 12667461.
- Lima WF, Rose JB, Nichols JG, et al. (2007). "Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate". Mol. Pharmacol. 71 (1): 83–91. doi:10.1124/mol.106.025015. PMID 17028158.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.