West Nile virus
| West Nile virus | |
|---|---|
 | |
| A micrograph of the West Nile Virus, appearing in yellow | |
| Virus classification | |
| Group: | Group IV ((+)ssRNA) |
| Family: | Flaviviridae |
| Genus: | Flavivirus |
| Species: | West Nile virus |
West Nile virus (WNV) is a single-stranded RNA virus that causes West Nile fever. It is a member of the family Flaviviridae, specifically from the genus Flavivirus, which also contains the Zika virus, dengue virus, and yellow fever virus. West Nile virus is primarily transmitted by mosquitoes, mostly species of the genus Culex, but ticks have also been found to carry the virus. The primary hosts of WNV are birds, so that the virus remains within a "bird-mosquito-bird" transmission cycle.[1]
Structure
Like most other flaviviruses, WNV is an enveloped virus, but has icosahedral symmetry.[2] Flavivirus envelopes consist of a protein shell and a lipid membrane. The protein shell is made of two structural proteins: the glycoprotein E and the small membrane protein M.[3] Protein E serve numerous functions, several of which include receptor binding, viral attachment, and entry into the cell through membrane fusion.[3][3]
The flavivirus lipid membrane has been found to contain cholesterol and phosphatidylserine, but other elements of the membrane have yet to be identified.[4][5] The lipid membrane has many roles in viral infection, including acting as signaling molecules and enhancing entry into the cell.[6] Cholesterol, in particular, plays an integral part in WNV entering a host cell.[7]
Within the viral envelope, the genome is contained within a protein capsid, which is one of the first proteins created in an infected cell. It has been found that the capsid prevents apoptosis by affecting the Akt pathway.[8] The capsid is a structural protein and its main purpose is to package RNA into the developing viruses.[9]
Genome
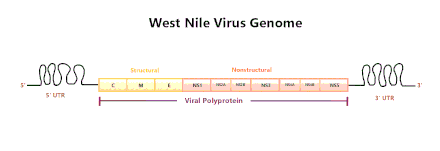
WNV is a positive-sense, single-stranded RNA virus. Its genome is approximately 11,000 nucleotides long and is flanked by 5' and 3' non-coding stem loop structures.[12] The coding region of the genome codes for three structural proteins and seven nonstructural (NS) proteins, proteins that are not incorporated into the structure of new viruses. The WNV genome is first translated into a polyprotein and later cleaved by virus and host proteases into separate proteins (i.e. NS1, C, E).[13]
Structural proteins
Structural proteins (C, prM/M, E) are capsid, precursor membrane proteins, and envelope proteins, respectively.[12] The structural proteins are located at the 5' end of the genome and are cleaved into mature proteins by proteases.
| Structural Protein | Function |
|---|---|
| C | Capsid protein; encloses the RNA genome, packages RNA into immature virions.[9][14] |
| prM/M | Viruses with M protein are infectious: the presence of M protein allows for the activation of proteins involved in viral entry into the cell. prM (precursor membrane) protein is present on immature virions, by further cleavage by furin to M protein, the virions become infectious.[15] |
| E | A glycoprotein that forms the viral envelope, binds to receptors on the host cell surface in order to enter the cell.[16] |
Nonstructural proteins
Nonstructural proteins consist of NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. These proteins mainly assist with viral replication or act as proteases.[14] The nonstructural proteins are located near the 3' end of the genome.
| Nonstructural Protein | Function |
|---|---|
| NS1 | NS1 is a cofactor for viral replication, specifically for regulation of the replication complex.[17] |
| NS2A | NS2A has a variety of functions: it is involved in viral replication, virion assembly, and inducing host cell death.[18] |
| NS2B | A cofactor for NS3 and together forms the NS2B-NS3 protease complex.[14] |
| NS3 | A protease that is responsible for cleaving the polyprotein to produce mature proteins; it can also acts as a helicase.[12] |
| NS4A | NS4A is a cofactor for viral replication, specifically regulates the activity of the NS3 helicase.[19] |
| NS4B | Inhibits interferon signaling.[20] |
| NS5 | The largest and most conserved protein of WNV, NS5 acts as a methyltransferase and a RNA polymerase, though it lacks proofreading properties.[14][21] |
Life cycle
Once WNV has successfully entered the bloodstream of a host animal, the envelope protein, E, binds to attachment factors called glycosaminoglycans on the host cell.[16] These attachment factors aid entry into the cell, however, binding to primary receptors is also necessary.[22] Primary receptors include DC-SIGN, DC-SIGN-R, and the integrin αvβ3.[23] By binding to these primary receptors, WNV enters the cell through clathrin-mediated endocytosis.[24] As a result of endocytosis, WNV enters the cell within an endosome.
The acidity of the endosome catalyzes the fusion of the endosomal and viral membranes, allowing the genome to be released into the cytoplasm.[25] Translation of the positive-sense single-stranded RNA occurs at the endoplasmic reticulum; the RNA is translated into a polyprotein which is then cleaved by viral proteases NS2B-N23 to produce mature proteins.[26]
In order to replicate its DNA, NS5, a RNA polymerase, forms a replication complex with other nonstructural proteins to produce an intermediary negative-sense single-stranded RNA; the negative-sense strand serves as a template for synthesis of the final positive-sense RNA.[22] Once the positive-sense RNA has been synthesized, the capsid protein, C, encloses the RNA strands into immature virions.[23] The rest of the virus is assembled along the endoplasmic reticulum and through the Golgi apparatus, and results in non-infectious immature virions.[26] The E protein is then glycosylated and prM is cleaved by furin, a host cell protease, into the M protein, thereby producing an infectious mature virion.[12][26] The mature viruses are then secreted out of the cell.
Phylogeny

Studies of phylogenetic lineages have determined that WNV emerged as a distinct virus around 1000 years ago.[28] This initial virus developed into two distinct lineages. Lineage 1 and its multiple profiles is the source of the epidemic transmission in Africa and throughout the world. Lineage 2 was considered an African zoonosis. However, in 2008, lineage 2, previously only seen in horses in sub-Saharan Africa and Madagascar, began to appear in horses in Europe, where the first known outbreak affected 18 animals in Hungary.[29] Lineage 1 West Nile virus was detected in South Africa in 2010 in a mare and her aborted fetus; previously, only lineage 2 West Nile virus had been detected in horses and humans in South Africa.[30] A 2007 fatal case in a killer whale in Texas broadened the known host range of West Nile virus to include cetaceans.[31]
Since the first North American cases in 1999, the virus has been reported throughout the United States, Canada, Mexico, the Caribbean, and Central America. There have been human cases and equine cases, and many birds are infected. The Barbary macaque, Macaca sylvanus, was the first nonhuman primate to contract WNV.[32] Both the American and Israeli strains are marked by high mortality rates in infected avian populations; the presence of dead birds—especially Corvidae—can be an early indicator of the arrival of the virus.
See also
References
- ↑ Mackenzie, John S; Gubler, Duane J; Petersen, Lyle R (2004). "Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses". Nature Medicine. 10 (12s): S98–S109. doi:10.1038/nm1144.
- ↑ Mukhopadhyay, Suchetana; Kim, Bong-Suk; Chipman, Paul R.; Rossmann, Michael G.; Kuhn, Richard J. (2003-10-10). "Structure of West Nile Virus". Science. 302 (5643): 248–248. doi:10.1126/science.1089316. ISSN 0036-8075. PMID 14551429.
- 1 2 3 Kanai, Ryuta; Kar, Kalipada; Anthony, Karen; Gould, L. Hannah; Ledizet, Michel; Fikrig, Erol; Marasco, Wayne A.; Koski, Raymond A.; Modis, Yorgo (2006-11-01). "Crystal Structure of West Nile Virus Envelope Glycoprotein Reveals Viral Surface Epitopes". Journal of Virology. 80 (22): 11000–11008. doi:10.1128/jvi.01735-06. ISSN 0022-538X. PMC 1642136. PMID 16943291.
- ↑ Meertens, Laurent; Carnec, Xavier; Lecoin, Manuel Perera; Ramdasi, Rasika; Guivel-Benhassine, Florence; Lew, Erin; Lemke, Greg; Schwartz, Olivier; Amara, Ali (2012). "The TIM and TAM Families of Phosphatidylserine Receptors Mediate Dengue Virus Entry". Cell Host & Microbe. 12 (4): 544–557. doi:10.1016/j.chom.2012.08.009.
- ↑ Carro, Ana C.; Damonte, Elsa B. (2013). "Requirement of cholesterol in the viral envelope for dengue virus infection". Virus Research. 174 (1–2): 78–87. doi:10.1016/j.virusres.2013.03.005.
- ↑ Martín-Acebes, Miguel A.; Merino-Ramos, Teresa; Blázquez, Ana-Belén; Casas, Josefina; Escribano-Romero, Estela; Sobrino, Francisco; Saiz, Juan-Carlos (2014-10-15). "The Composition of West Nile Virus Lipid Envelope Unveils a Role of Sphingolipid Metabolism in Flavivirus Biogenesis". Journal of Virology. 88 (20): 12041–12054. doi:10.1128/jvi.02061-14. ISSN 0022-538X. PMC 4178726. PMID 25122799.
- ↑ Medigeshi, Guruprasad R.; Hirsch, Alec J.; Streblow, Daniel N.; Nikolich-Zugich, Janko; Nelson, Jay A. (2008-06-01). "West Nile Virus Entry Requires Cholesterol-Rich Membrane Microdomains and Is Independent of αvβ3 Integrin". Journal of Virology. 82 (11): 5212–5219. doi:10.1128/jvi.00008-08. ISSN 0022-538X. PMC 2395215. PMID 18385233.
- ↑ Urbanowski, Matt D.; Hobman, Tom C. (2013-01-15). "The West Nile Virus Capsid Protein Blocks Apoptosis through a Phosphatidylinositol 3-Kinase-Dependent Mechanism". Journal of Virology. 87 (2): 872–881. doi:10.1128/jvi.02030-12. ISSN 0022-538X. PMC 3554064. PMID 23115297.
- 1 2 Hunt, Tracey A.; Urbanowski, Matthew D.; Kakani, Kishore; Law, Lok-Man J.; Brinton, Margo A.; Hobman, Tom C. (2007-11-01). "Interactions between the West Nile virus capsid protein and the host cell-encoded phosphatase inhibitor, I2PP2A". Cellular Microbiology. 9 (11): 2756–2766. doi:10.1111/j.1462-5822.2007.01046.x. ISSN 1462-5822.
- ↑ Guzman, Maria G.; Halstead, Scott B.; Artsob, Harvey; Buchy, Philippe; Farrar, Jeremy; Gubler, Duane J.; Hunsperger, Elizabeth; Kroeger, Axel; Margolis, Harold S. (2010-12-01). "Dengue: a continuing global threat". Nature Reviews Microbiology. 8 (12): S7–S16. doi:10.1038/nrmicro2460. ISSN 1740-1534.
- ↑ "Dengue Viruses". www.nature.com. Retrieved 2017-12-19.
- 1 2 3 4 Colpitts, Tonya M.; Conway, Michael J.; Montgomery, Ruth R.; Fikrig, Erol (2012-10-01). "West Nile Virus: Biology, Transmission, and Human Infection". Clinical Microbiology Reviews. 25 (4): 635–648. doi:10.1128/cmr.00045-12. ISSN 0893-8512. PMC 3485754. PMID 23034323.
- ↑ Chung, Kyung Min; Liszewski, M. Kathryn; Nybakken, Grant; Davis, Alan E.; Townsend, R. Reid; Fremont, Daved H.; Atkinson, John P.; Diamond, Michael S. (2006-12-12). "West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H". Proceedings of the National Academy of Sciences. 103 (50): 19111–19116. doi:10.1073/pnas.0605668103. ISSN 0027-8424. PMC 1664712. PMID 17132743.
- 1 2 3 4 Londono-Renteria, Berlin; Colpitts, Tonya M. (2016). "A Brief Review of West Nile Virus Biology". Methods in Molecular Biology (Clifton, N.J.). 1435: 1–13. doi:10.1007/978-1-4939-3670-0_1. ISSN 1940-6029. PMID 27188545.
- ↑ Moesker, Bastiaan; Rodenhuis-Zybert, Izabela A.; Meijerhof, Tjarko; Wilschut, Jan; Smit, Jolanda M. (2010). "Characterization of the functional requirements of West Nile virus membrane fusion". Journal of General Virology. 91 (2): 389–393. doi:10.1099/vir.0.015255-0.
- 1 2 Perera-Lecoin, Manuel; Meertens, Laurent; Carnec, Xavier; Amara, Ali (2013-12-30). "Flavivirus Entry Receptors: An Update". Viruses. 6 (1): 69–88. doi:10.3390/v6010069.
- ↑ Youn, Soonjeon; Ambrose, Rebecca L.; Mackenzie, Jason M.; Diamond, Michael S. (2013-11-18). "Non-structural protein-1 is required for West Nile virus replication complex formation and viral RNA synthesis". Virology Journal. 10: 339. doi:10.1186/1743-422x-10-339. ISSN 1743-422X.
- ↑ Melian, Ezequiel Balmori; Edmonds, Judith H.; Nagasaki, Tomoko Kim; Hinzman, Edward; Floden, Nadia; Khromykh, Alexander A. (2013). "West Nile virus NS2A protein facilitates virus-induced apoptosis independently of interferon response". Journal of General Virology. 94 (2): 308–313. doi:10.1099/vir.0.047076-0. PMC 3709616.
- ↑ Shiryaev, Sergey A.; Chernov, Andrei V.; Aleshin, Alexander E.; Shiryaeva, Tatiana N.; Strongin, Alex Y. (2009). "NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus". Journal of General Virology. 90 (9): 2081–2085. doi:10.1099/vir.0.012864-0. PMC 2887571.
- ↑ Wicker, Jason A.; Whiteman, Melissa C.; Beasley, David W.C.; Davis, C. Todd; McGee, Charles E.; Lee, J. Ching; Higgs, Stephen; Kinney, Richard M.; Huang, Claire Y.-H. (2012). "Mutational analysis of the West Nile virus NS4B protein". Virology. 426 (1): 22–33. doi:10.1016/j.virol.2011.11.022.
- ↑ Davidson, Andrew D. Chapter 2 New Insights into Flavivirus Nonstructural Protein 5. pp. 41–101. doi:10.1016/s0065-3527(09)74002-3.
- 1 2 Brinton, Margo A. (2002-10-01). "The Molecular Biology of West Nile Virus: A New Invader of the Western Hemisphere". Annual Review of Microbiology. 56 (1): 371–402. doi:10.1146/annurev.micro.56.012302.160654. ISSN 0066-4227.
- 1 2 Samuel, Melanie A.; Diamond, Michael S. (2006-10-01). "Pathogenesis of West Nile Virus Infection: a Balance between Virulence, Innate and Adaptive Immunity, and Viral Evasion". Journal of Virology. 80 (19): 9349–9360. doi:10.1128/jvi.01122-06. ISSN 0022-538X. PMC 1617273. PMID 16973541.
- ↑ Vancini, Ricardo; Kramer, Laura D.; Ribeiro, Mariana; Hernandez, Raquel; Brown, Dennis (2013). "Flavivirus infection from mosquitoes in vitro reveals cell entry at the plasma membrane". Virology. 435 (2): 406–414. doi:10.1016/j.virol.2012.10.013.
- ↑ Mukhopadhyay, Suchetana; Kuhn, Richard J.; Rossmann, Michael G. (2005). "A structural perspective of the flavivirus life cycle". Nature Reviews Microbiology. 3 (1): 13–22. doi:10.1038/nrmicro1067.
- 1 2 3 Suthar, Mehul S.; Diamond, Michael S.; Jr, Michael Gale (2013). "West Nile virus infection and immunity". Nature Reviews Microbiology. 11 (2): 115–128. doi:10.1038/nrmicro2950.
- ↑ Lanciotti RS, Ebel GD, Deubel V, et al. (June 2002). "Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East". Virology. 298 (1): 96–105. doi:10.1006/viro.2002.1449. PMID 12093177.
- ↑ Galli M, Bernini F, Zehender G (July 2004). "Alexander the Great and West Nile virus encephalitis". Emerging Infect. Dis. 10 (7): 1330–2, author reply 1332–3. doi:10.3201/eid1007.040396. PMID 15338540.
- ↑ West, Christy (2010-02-08). "Different West Nile Virus Genetic Lineage Evolving?". The Horse. Archived from the original on 2010-02-17. Retrieved 2010-02-10. From statements by Orsolya Kutasi, DVM, of the Szent Istvan University, Hungary at the 2009 American Association of Equine Practitioners Convention, December 5–9, 2009.
- ↑ Venter M, Human S, van Niekerk S, Williams J, van Eeden C, Freeman F (August 2011). "Fatal neurologic disease and abortion in mare infected with lineage 1 West Nile virus, South Africa". Emerging Infect. Dis. 17 (8): 1534–6. doi:10.3201/eid1708.101794. PMC 3381566. PMID 21801644.
- ↑ St Leger J, Wu G, Anderson M, Dalton L, Nilson E, Wang D (2011). "West Nile virus infection in killer whale, Texas, USA, 2007". Emerging Infect. Dis. 17 (8): 1531–3. doi:10.3201/eid1708.101979. PMC 3381582. PMID 21801643.
- ↑ Hogan, C. Michael (2008). Barbary Macaque: Macaca sylvanus, GlobalTwitcher.com Archived 2009-08-31 at the Wayback Machine.