25-Hydroxyvitamin D3 1-alpha-hydroxylase
25-Hydroxyvitamin D3 1-alpha-hydroxylase (VD3 1A hydroxylase) also known as cytochrome p450 27B1 (CYP27B1) or simply 1-alpha-hydroxylase is a cytochrome P450 enzyme that in humans is encoded by the CYP27B1 gene.[5][6][7]
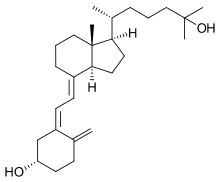
VD3 1A hydroxylase is located in the proximal tubule of the kidney and a variety of other tissues, including skin (keratinocytes), immune cells,[8] and bone (osteoblasts).[9] The enzyme catalyzes the hydroxylation of Calcifediol to calcitriol (the bioactive form of Vitamin D):[10]
- calcidiol + 2 reduced adrenodoxin + 2 H+ + O2 ⇌ calcitriol + 2 oxidized adrenodoxin + H2O
| ||||||||||||||||||||||||||||||||||||
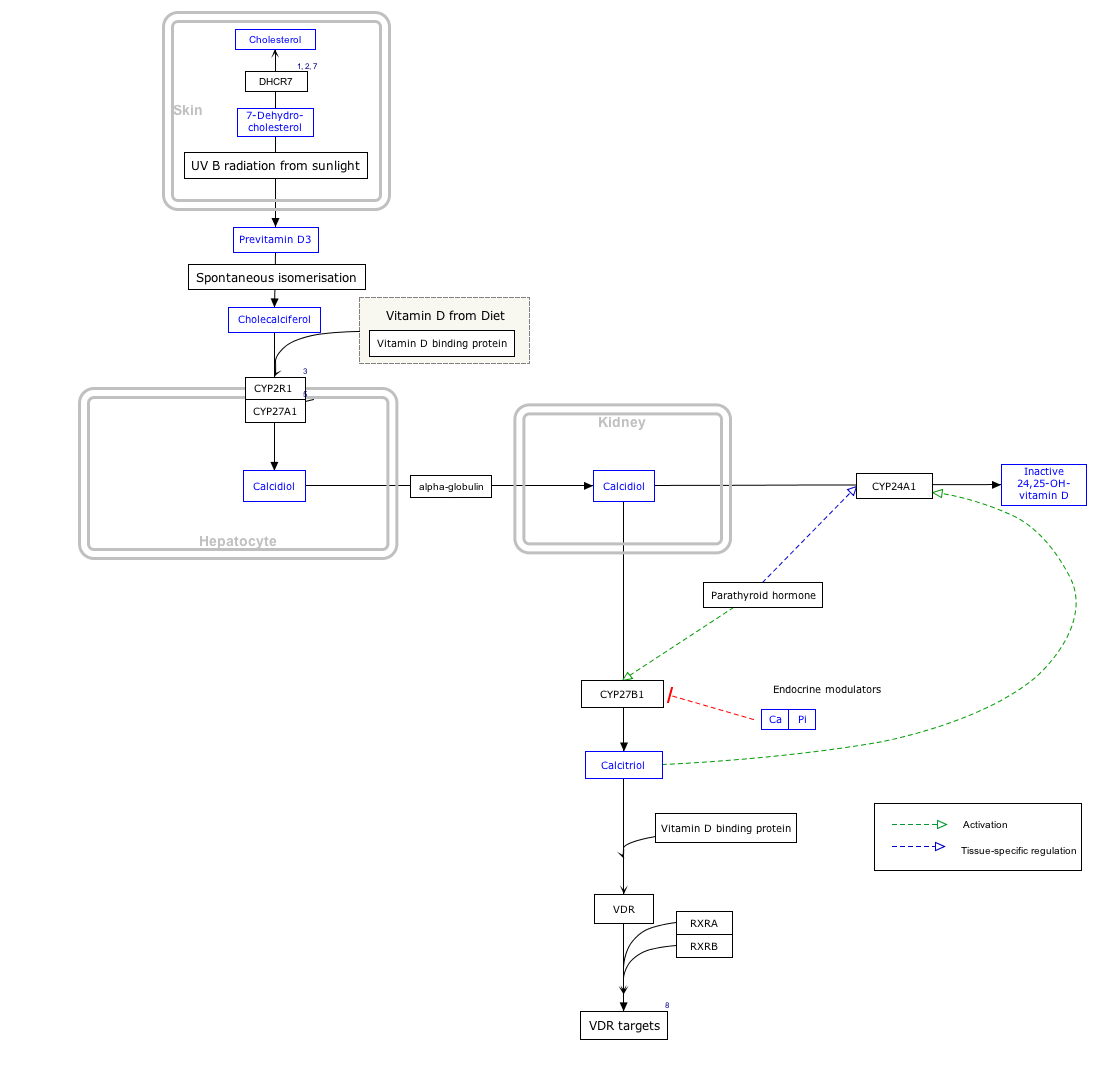
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ↑ The interactive pathway map can be edited at WikiPathways: "VitaminDSynthesis_WP1531".
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000111012 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000006724 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ "Entrez Gene: cytochrome P450".
- ↑ Takeyama K, Kitanaka S, Sato T, Kobori M, Yanagisawa J, Kato S (Sep 1997). "25-Hydroxyvitamin D3 1alpha-hydroxylase and vitamin D synthesis". Science. 277 (5333): 1827–30. doi:10.1126/science.277.5333.1827. PMID 9295274.
- ↑ Monkawa T, Yoshida T, Wakino S, Shinki T, Anazawa H, Deluca HF, Suda T, Hayashi M, Saruta T (Oct 1997). "Molecular cloning of cDNA and genomic DNA for human 25-hydroxyvitamin D3 1 alpha-hydroxylase". Biochemical and Biophysical Research Communications. 239 (2): 527–33. doi:10.1006/bbrc.1997.7508. PMID 9344864.
- ↑ Sigmundsdottir H, Pan J, Debes GF, Alt C, Habtezion A, Soler D, Butcher EC (Mar 2007). "DCs metabolize sunlight-induced vitamin D3 to 'program' T cell attraction to the epidermal chemokine CCL27" (PDF). Nature Immunology. 8 (3): 285–93. doi:10.1038/ni1433. PMID 17259988.
- ↑ Kogawa M, Findlay DM, Anderson PH, Ormsby R, Vincent C, Morris HA, Atkins GJ (Oct 2010). "Osteoclastic metabolism of 25(OH)-vitamin D3: a potential mechanism for optimization of bone resorption". Endocrinology. 151 (10): 4613–25. doi:10.1210/en.2010-0334. PMID 20739402.
- ↑ Gray RW, Omdahl JL, Ghazarian JG, DeLuca HF (Dec 1972). "25-Hydroxycholecalciferol-1-hydroxylase. Subcellular location and properties". The Journal of Biological Chemistry. 247 (23): 7528–32. PMID 4404596.
Further reading
- Carr EJ, Niederer HA, Williams J, Harper L, Watts RA, Lyons PA, Smith KG (2009). "Confirmation of the genetic association of CTLA4 and PTPN22 with ANCA-associated vasculitis". BMC Medical Genetics. 10: 121. doi:10.1186/1471-2350-10-121. PMID 19951419.
- Alzahrani AS, Zou M, Baitei EY, Alshaikh OM, Al-Rijjal RA, Meyer BF, Shi Y (Sep 2010). "A novel G102E mutation of CYP27B1 in a large family with vitamin D-dependent rickets type 1". The Journal of Clinical Endocrinology and Metabolism. 95 (9): 4176–83. doi:10.1210/jc.2009-2278. PMID 20534770.
- Lagishetty V, Chun RF, Liu NQ, Lisse TS, Adams JS, Hewison M (Jul 2010). "1alpha-hydroxylase and innate immune responses to 25-hydroxyvitamin D in colonic cell lines". The Journal of Steroid Biochemistry and Molecular Biology. 121 (1–2): 228–33. doi:10.1016/j.jsbmb.2010.02.004. PMC 2891066. PMID 20152900.
- Giroux S, Elfassihi L, Clément V, Bussières J, Bureau A, Cole DE, Rousseau F (Nov 2010). "High-density polymorphisms analysis of 23 candidate genes for association with bone mineral density". Bone. 47 (5): 975–81. doi:10.1016/j.bone.2010.06.030. PMID 20654748.
- Zhou S, LeBoff MS, Glowacki J (Jan 2010). "Vitamin D metabolism and action in human bone marrow stromal cells". Endocrinology. 151 (1): 14–22. doi:10.1210/en.2009-0969. PMC 2803155. PMID 19966181.
- Payne AH, Hales DB (Dec 2004). "Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones". Endocrine Reviews. 25 (6): 947–70. doi:10.1210/er.2003-0030. PMID 15583024.
- Maver A, Medica I, Salobir B, Tercelj M, Peterlin B (2010). "Lack of association of immune-response-gene polymorphisms with susceptibility to sarcoidosis in Slovenian patients". Genetics and Molecular Research. 9 (1): 58–68. doi:10.4238/vol9-1gmr682. PMID 20082271.
- Shen H, Bielak LF, Ferguson JF, Streeten EA, Yerges-Armstrong LM, Liu J, Post W, O'Connell JR, Hixson JE, Kardia SL, Sun YV, Jhun MA, Wang X, Mehta NN, Li M, Koller DL, Hakonarson H, Keating BJ, Rader DJ, Shuldiner AR, Peyser PA, Reilly MP, Mitchell BD (Dec 2010). "Association of the vitamin D metabolism gene CYP24A1 with coronary artery calcification". Arteriosclerosis, Thrombosis, and Vascular Biology. 30 (12): 2648–54. doi:10.1161/ATVBAHA.110.211805. PMC 2988112. PMID 20847308.
- Sundqvist E, Bäärnhielm M, Alfredsson L, Hillert J, Olsson T, Kockum I (Dec 2010). "Confirmation of association between multiple sclerosis and CYP27B1". European Journal of Human Genetics. 18 (12): 1349–52. doi:10.1038/ejhg.2010.113. PMC 3002863. PMID 20648053.
- Fichna M, Zurawek M, Januszkiewicz-Lewandowska D, Gryczyñska M, Fichna P, Sowiñski J, Nowak J (Aug 2010). "Association of the CYP27B1 C(-1260)A polymorphism with autoimmune Addison's disease". Experimental and Clinical Endocrinology & Diabetes. 118 (8): 544–9. doi:10.1055/s-0029-1241206. PMID 19998245.
- Holt SK, Kwon EM, Koopmeiners JS, Lin DW, Feng Z, Ostrander EA, Peters U, Stanford JL (Sep 2010). "Vitamin D pathway gene variants and prostate cancer prognosis". The Prostate. 70 (13): 1448–60. doi:10.1002/pros.21180. PMC 2927712. PMID 20687218.
- Bu FX, Armas L, Lappe J, Zhou Y, Gao G, Wang HW, Recker R, Zhao LJ (Nov 2010). "Comprehensive association analysis of nine candidate genes with serum 25-hydroxy vitamin D levels among healthy Caucasian subjects". Human Genetics. 128 (5): 549–56. doi:10.1007/s00439-010-0881-9. PMID 20809279.
- Fichna M, Zurawek M, Januszkiewicz-Lewandowska D, Fichna P, Nowak J (Oct 2010). "PTPN22, PDCD1 and CYP27B1 polymorphisms and susceptibility to type 1 diabetes in Polish patients". International Journal of Immunogenetics. 37 (5): 367–72. doi:10.1111/j.1744-313X.2010.00935.x. PMID 20518841.
- Wjst M, Heimbeck I, Kutschke D, Pukelsheim K (Jul 2010). "Epigenetic regulation of vitamin D converting enzymes". The Journal of Steroid Biochemistry and Molecular Biology. 121 (1–2): 80–3. doi:10.1016/j.jsbmb.2010.03.056. PMID 20304056.
- Liu CY, Wu MC, Chen F, Ter-Minassian M, Asomaning K, Zhai R, Wang Z, Su L, Heist RS, Kulke MH, Lin X, Liu G, Christiani DC (Jul 2010). "A Large-scale genetic association study of esophageal adenocarcinoma risk". Carcinogenesis. 31 (7): 1259–63. doi:10.1093/carcin/bgq092. PMC 2893800. PMID 20453000.
- Sunyer J, Basagaña X, González JR, Júlvez J, Guerra S, Bustamante M, de Cid R, Antó JM, Torrent M (Oct 2010). "Early life environment, neurodevelopment and the interrelation with atopy". Environmental Research. 110 (7): 733–8. doi:10.1016/j.envres.2010.07.005. PMID 20701904.
- Simon KC, Munger KL, Ascherio A (Feb 2010). "Polymorphisms in vitamin D metabolism related genes and risk of multiple sclerosis". Multiple Sclerosis (Houndmills, Basingstoke, England). 16 (2): 133–8. doi:10.1177/1352458509355069. PMC 2819633. PMID 20007432.
- Hendrickson SL, Lautenberger JA, Chinn LW, Malasky M, Sezgin E, Kingsley LA, Goedert JJ, Kirk GD, Gomperts ED, Buchbinder SP, Troyer JL, O'Brien SJ (2010). Badger JH, ed. "Genetic variants in nuclear-encoded mitochondrial genes influence AIDS progression". PLOS ONE. 5 (9): e12862. doi:10.1371/journal.pone.0012862. PMC 2943476. PMID 20877624.
- Dusso AS, Brown AJ, Slatopolsky E (Jul 2005). "Vitamin D". American Journal of Physiology. Renal Physiology. 289 (1): F8–28. doi:10.1152/ajprenal.00336.2004. PMID 15951480.
- Bailey SD, Xie C, Do R, Montpetit A, Diaz R, Mohan V, Keavney B, Yusuf S, Gerstein HC, Engert JC, Anand S (Oct 2010). "Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study". Diabetes Care. 33 (10): 2250–3. doi:10.2337/dc10-0452. PMC 2945168. PMID 20628086.
External links
- 25-Hydroxyvitamin+D3+1-alpha-Hydroxylase at the US National Library of Medicine Medical Subject Headings (MeSH)
- Human CYP27B1 genome location and CYP27B1 gene details page in the UCSC Genome Browser.
- Human VDR genome location and VDR gene details page in the UCSC Genome Browser.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.