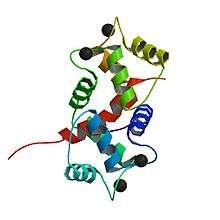
Calmodulin
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells.[1] It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases.[2][3][4]
| Calmodulin | |
|---|---|
 | |
| Identifiers | |
| Symbol | CaM |
| PDB | 1OSA |
| UniProt | P62158 |
Structure
Calmodulin is a small, highly conserved protein that is 148 amino acids long (16.7 KDa). The protein has two approximately symmetrical globular domains each containing a pair of EF hand motifs (the N- and C-domain) separated by a flexible linker region for a total of four Ca2+ binding sites.[5] Each hand motif allows calmodulin to sense intracellular calcium levels by binding one Ca2+ ion. Calcium ion binding regions are found in the following positions in the sequence of amino acids: 21–32, 57–68, 94–105 and 130–141; each region that calcium binds to is exactly 12 amino acids long. These regions are located between two alpha helices in the EF-hand motifs, the first two regions (21–32 and 57–68) are on one side of the linker region the other two (94–105 and 130–141) are on the other side.[4]
Importance of flexibility in calmodulin
Calmodulin binds such a wide variety of target proteins, making it especially important for it to have flexibility. Though calmodulin's flexibility is more evident when it is bound to a target protein, NMR studies have shown that the linker region of calmodulin is flexible, even when it is not bound to a target protein. Another important characteristic of calmodulin that allows it to bind a large variety of target proteins is the generic shape of the non-polar grooves in the binding sites. Since the non-polar grooves are generic, they don't require the target proteins to have any specific sequence of amino acids allowing a larger variety of target proteins to be bound. Together, these two structural characteristics of calmodulin allow it to flexibly bind target proteins with various shapes and amino acid sequences.[5] For example, calmodulin binds both NMDA receptors and potassium channels which differ in length by about 50 amino acid residues.[6][7]
Similarity to troponin C
Calmodulin's structure is very similar to the structure of troponin C (which is another calcium binding protein). They are both members of the EFh superfamily. Troponin C, like calmodulin, has two globular domains that are connected by a linker region.[8] However, troponin C and calmodulin differ in the length of the linker region; the linker region of calmodulin is smaller than that of troponin C.[8] These remarkably similar structures are an example of how the EF hand motif is highly conserved in calcium binding proteins. Though they have similar structures, their functions are very different. Troponin C has a very specific function (to elicit a conformational change in troponin I) ultimately causing a contraction in skeletal muscles. Calmodulin evolved to bind a wider variety of target proteins, allowing it to play a role in many physiological events.[5][8]
Mechanism
Overall
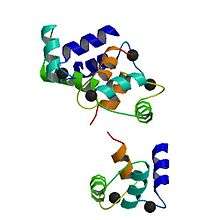
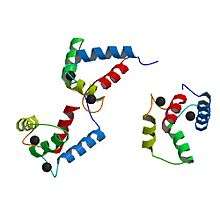
Up to four calcium ions are bound by calmodulin via its four EF hand motifs.[5] EF hands supply an electronegative environment for ion coordination. After calcium binding, hydrophobic methyl groups from methionine residues become exposed on the protein via conformational change. Using both X-ray spectroscopy and NMR studies, scientists were able to determine that the conformational changes occurred in the alpha-helices of the EF motif, which changes the binding affinity for target proteins. When the alpha helices are perpendicular to one another, the Calmodulin is in an open conformation giving it a higher affinity for target proteins.[9] More specifically, this conformational change presents hydrophobic surfaces, which can in turn bind to Basic Amphiphilic Helices (BAA helices) on the target protein. These helices contain complementary hydrophobic regions. The flexibility of calmodulin's hinged region allows the molecule to wrap around its target.[5] This property allows it to tightly bind to a wide range of different target proteins. The C-domain of calmodulin has a higher affinity for calcium than does the N-domain.
Dynamic features
The C-terminal domain solution structure is similar to the X-ray crystal structure, while the EF hands of the N-terminal domain are considerably less open to the X-ray crystal structure. This indicates that Ca2+ binding causes a larger conformational change in the N-domain than in the C-domain. The backbone flexibility within calmodulin is key to its ability to bind a wide range of targets.[10] Protein domains, connected by intrinsically disordered flexible linker domains, induce long-range allostery, or the conformational change of a protein by ligand binding to an allosteric site (a site other than the functional site), due to protein domain dynamics.[10]
Target binding and recognition
Binding of calcium ions causes large conformational changes in CaM, which further selectively binds and activates downstream CaM binding target proteins (CaMBTs), in addition to those independent of Ca2+.[1] There are several opinions of the binding mechanism between CaM and CaMBT. The binding between a CaM and a CaMBT involves conformational changes in both. The binding domain of the CaMBTs is usually disordered. Current research indicates that selective protein binding occurs through the mechanism of mutually induced conformational fit,[11] which would explain how calcium dynamics in CaM would modulate its interaction.
Current research on CaM signaling and CaM–BT interaction includes experimental kinetic rate observations and coarse grain/all atom molecular dynamics simulations. Because protein signaling and protein–protein interaction is a new field of research, many observed interactions cannot be explained through experiment alone. The unification between simulation and experimental results is necessary to expand the predictive power of the theoretical approach and create general laws that explain the mechanics of signaling/protein–protein interactions.
The computational approach for modeling macro molecules is very resource intensive. The Hamiltonian equation in molecular dynamic software relates each atom to all other atoms in the system through kinetic, electrostatic, van der Waals, dihedral angle, bond, etc. energies. For example, calmodulin-binding domain of brain calmodulin-dependent protein kinase II alpha polypeptide contains 21 residues and 318 atoms. For a single time step, the molecular dynamics software must perform energy calculations between every atom in the polypeptide, which is ~100,000 calculations. Since the time step must be in the sub picosecond range (to insure stability), several million time steps must be performed to obtain meaningful data. To remedy the large number of calculations involved in all atom simulations, the coarse grain simulation technique can be used. Current work from the biophysics group at the University of Houston uses open source coarse-grained and all atomistic models of CaM and wildtype/mutated binding targets of CaMKII in their research.
Role in animals
Calmodulin mediates many crucial processes such as inflammation, metabolism, apoptosis, smooth muscle contraction, intracellular movement, short-term and long-term memory, and the immune response.[12][13] Calcium participates in an intracellular signaling system by acting as a diffusible second messenger to the initial stimuli. It does this by binding various targets in the cell including a large number of enzymes, ion channels, aquaporins and other proteins.[4] Calmodulin is expressed in many cell types and can have different subcellular locations, including the cytoplasm, within organelles, or associated with the plasma or organelle membranes, but it is always found intracellularly.[13] Many of the proteins that calmodulin binds are unable to bind calcium themselves, and use calmodulin as a calcium sensor and signal transducer. Calmodulin can also make use of the calcium stores in the endoplasmic reticulum, and the sarcoplasmic reticulum. Calmodulin can undergo post-translational modifications, such as phosphorylation, acetylation, methylation and proteolytic cleavage, each of which has potential to modulate its actions.
Specific examples
Role in smooth muscle contraction
Calmodulin plays an important role in excitation contraction (EC) coupling and the initiation of the cross-bridge cycling in smooth muscle, ultimately causing smooth muscle contraction.[14] In order to activate contraction of smooth muscle, the head of the myosin light chain must be phosphorylated. This phosphorylation is done by myosin light chain (MLC) kinase. This MLC kinase is activated by a calmodulin when it is bound by calcium, thus making smooth muscle contraction dependent on the presence of calcium, through the binding of calmodulin and activation of MLC kinase.[14]
Another way that calmodulin affects muscle contraction is by controlling the movement of Ca2+ across both the cell and sarcoplasmic reticulum membranes. The Ca2+ channels, such as the ryanodine receptor of the sarcoplasmic reticulum, can be inhibited by calmodulin bound to calcium, thus affecting the overall levels of calcium in the cell.[15] Calcium pumps take calcium out of the cytoplasm or store it in the endoplasmic reticulum and this control helps regulate many downstream processes.
This is a very important function of calmodulin because it indirectly plays a role in every physiological process that is affected by smooth muscle contraction such as digestion and contraction of arteries (which helps distribute blood and regulate blood pressure).[16]
Role in metabolism
Calmodulin plays an important role in the activation of phosphorylase kinase, which ultimately leads to glucose being cleaved from glycogen by glycogen phosphorylase.[17]
Calmodulin also plays an important role in lipid metabolism by affecting Calcitonin. Calcitonin is a polypeptide hormone that lowers blood Ca2+ levels and activates G protein cascades that leads to the generation of cAMP. The actions of calcitonin can be blocked by inhibiting the actions of calmodulin, suggesting that calmodulin plays a crucial role in the activation of calcitonin.[17]
Role in short-term and long-term memory
Ca2+/calmodulin-dependent protein kinase II (CaMKII) plays a crucial role in a type of synaptic plasticity known as long-term potentiation (LTP) which requires the presence of calcium/calmodulin. CaMKII contributes to the phosphorylation of an AMPA receptor which increases the sensitivity of AMPA receptors.[18] Furthermore, research shows that inhibiting CaMKII interferes with LTP.[18]
Role in plants
.jpg)
While yeasts have only a single CaM gene, plants and vertebrates contain an evolutionarily conserved form of CaM genes. The difference between plants and animals in Ca2+ signaling is that the plants contain an extended family of the CaM in addition to the evolutionarily conserved form.[19] Calmodulins play an essential role in plant development and adaptation to environmental stimuli.
Calcium plays a key role in the structural integrity of the cell wall and the membrane system of the cell. However, high calcium levels can be toxic to a plant's cellular energy metabolism and, hence, the Ca2+ concentration in the cytosol is maintained at a submicromolar level by removing the cytosolic Ca2+ to either the apoplast or the lumen of the intracellular organelles. Ca2+ pulses created due to increased influx and efflux act as cellular signals in response to external stimuli such as hormones, light, gravity, abiotic stress factors and also interactions with pathogens.
CMLs (CaM-related proteins)
Plants contain CaM-related proteins (CMLs) apart from the typical CaM proteins. The CMLs have about 15% amino acid similarity with the typical CaMs. Arabidopsis thaliana contains about 50 different CML genes which leads to the question of what purpose these diverse ranges of proteins serve in the cellular function. All plant species exhibit this diversity in the CML genes. The different CaMs and CMLs differ in their affinity to bind and activate the CaM-regulated enzymes in vivo. The CaM or CMLs are also found to be located in different organelle compartments.
Plant growth and development
In Arabidopsis, the protein DWF1 plays an enzymatic role in the biosynthesis of brassinosteroids, steroid hormones in plants that are required for growth. An interaction occurs between CaM and DWF1, and DWF1 being unable to bind CaM is unable to produce a regular growth phenotype in plants. Hence, CaM is essential for the DWF1 function in plant growth.
CaM binding proteins are also known to regulate reproductive development in plants. For instance, the CaM-binding protein kinase in tobacco acts as a negative regulator of flowering. However, these CaM-binding protein kinase are also present in the shoot apical meristem of tobacco and a high concentration of these kinases in the meristem causes a delayed transition to flowering in the plant.
S-locus receptor kinase (SRK) is another protein kinase that interacts with CaM. SRK is involved in the self-incompatibility responses involved in pollen-pistil interactions in Brassica.
CaM targets in Arabidopsis are also involved in pollen development and fertilization. Ca2+ transporters are essential for pollen tube growth. Hence, a constant Ca2+ gradient is maintained at the apex of pollen tube for elongation during the process of fertilization. Similarly, CaM is also essential at the pollen tube apex, where its primarily role involves the guidance of the pollen tube growth.
Interaction with microbes
Nodule formation
Ca2+ plays a significantly important role in nodule formation in legumes. Nitrogen is an essential element required in plants and many legumes, unable to fix nitrogen independently, pair symbiotically with nitrogen-fixing bacteria that reduce nitrogen to ammonia. This legume-Rhizobium interaction establishment requires the Nod factor that is produced by the Rhizobium bacteria. The Nod factor is recognized by the root hair cells that are involved in the nodule formation in legumes. Ca2+ responses of varied nature are characterized to be involved in the Nod factor recognition. There is a Ca2+ flux at the tip of the root hair initially followed by repetitive oscillation of Ca2+ in the cytosol and also Ca2+ spike occurs around the nucleus. DMI3, an essential gene for Nod factor signaling functions downstream of the Ca2+ spiking signature, might be recognizing the Ca2+ signature. Further, several CaM and CML genes in Medicago and Lotus are expressed in nodules.
Pathogen defense
Among the diverse range of defense strategies plants utilize against pathogens, Ca2+ signaling is very increasingly common. Free Ca2+ levels in the cytoplasm increases in response to a pathogenic infection. Ca2+ signatures of this nature usually activate the plant defense system by inducing defense-related genes and the hypersensitive cell death. CaMs, CMLs and CaM-binding proteins are some of the recently identified elements of the plant defense signaling pathways. Several CML genes in tobacco, bean and tomato are responsive to pathogens. CML43 is a CaM-related protein that, as isolated from APR134 gene in the disease-resistant leaves of Arabidopsis for gene expression analysis, is rapidly induced when the leaves are inoculated with Pseudomonas syringae. These genes are also found in tomatoes (Solanum lycopersicum). The CML43 from the APR134 also binds to Ca2+ ions in vitro which shows that CML43 and APR134 are, hence, involved in the Ca2+-dependent signaling during the plant immune response to bacterial pathogens.[20] The CML9 expression in Arabidopsis thaliana is rapidly induced by phytopathogenic bacteria, flagellin and salicylic acid.[21] Expression of soybean SCaM4 and SCaM5 in transgenic tobacco and Arabidopsis causes an activation of genes related to pathogen resistance and also results in enhanced resistance to a wide spectrum of pathogen infection. The same is not true for soybean SCaM1 and SCaM2 that are highly conserved CaM isoforms. The AtBAG6 protein is a CaM-binding protein that binds to CaM only in the absence of Ca2+ and not in the presence of it. AtBAG6 is responsible for the hypersensitive response of programmed cell death in order to prevent the spread of pathogen infection or to restrict pathogen growth. Mutations in the CaM binding proteins can lead to severe effects on the defense response of the plants towards pathogen infections. Cyclic nucleotide-gated channels (CNGCs) are functional protein channels in the plasma membrane that have overlapping CaM binding sites transport divalent cations such as Ca2+. However, the exact role of the positioning of the CNGCs in this pathway for plant defense is still unclear.
Abiotic stress response in plants
Change in intracellular Ca2+ levels is used as a signature for diverse responses towards mechanical stimuli, osmotic and salt treatments, and cold and heat shocks. Different root cell types show a different Ca2+ response to osmotic and salt stresses and this implies the cellular specificities of Ca2+ patterns. In response to external stress CaM activates glutamate decarboxylase (GAD) that catalyzes the conversion of L-glutamate to GABA. A tight control on the GABA synthesis is important for plant development and, hence, increased GABA levels can essentially affect plant development. Therefore, external stress can affect plant growth and development and CaM are involved in that pathway controlling this effect.
Plant examples
Sorghum
The plant sorghum is well established model organism and can adapt in hot and dry environments. For this reason, it is used as a model to study calmodulin's role in plants.[22] Sorghum contains seedlings that express a glycine-rich RNA-binding protein, SbGRBP. This particular protein can be modulated by using heat as a stressor. Its unique location in the cell nucleus and cytosol demonstrates interaction with calmodulin that requires the use of Ca2+.[23] By exposing the plant to versatile stress conditions, it can cause different proteins that enable the plant cells to tolerate environmental changes to become repressed. These modulated stress proteins are shown to interact with CaM. The CaMBP genes expressed in the sorghum are depicted as a “model crop” for researching the tolerance to heat and drought stress.
Arabidopsis
In an Arabidopsis thaliana study, hundreds of different proteins demonstrated the possibility to bind to CaM in plants.[22]
Family members
- Calmodulin 1 (CALM1)
- Calmodulin 2 (CALM2)
- Calmodulin 3 (CALM3)
- calmodulin 1 pseudogene 1 (CALM1P1)
- Calmodulin-like 3 (CALML3)
- Calmodulin-like 4 (CALML4)
- Calmodulin-like 5 (CALML5)
- Calmodulin-like 6 (CALML6)
Other calcium-binding proteins
Calmodulin belongs to one of the two main groups of calcium-binding proteins, called EF hand proteins. The other group, called annexins, bind calcium and phospholipids such as lipocortin. Many other proteins bind calcium, although binding calcium may not be considered their principal function in the cell.
See also
- Proteopedia page for Calmodulin and its conformational change
- Protein kinase
- Ca2+/calmodulin-dependent protein kinase
References
- Stevens FC (August 1983). "Calmodulin: an introduction". Canadian Journal of Biochemistry and Cell Biology. 61 (8): 906–10. doi:10.1139/o83-115. PMID 6313166.
- Chin D, Means AR (August 2000). "Calmodulin: a prototypical calcium sensor". Trends in Cell Biology. 10 (8): 322–8. doi:10.1016/S0962-8924(00)01800-6. PMID 10884684.
- Purves D, Augustine G, Fitzpatrick D, Hall W, LaMantia A, White L (2012). Neuroscience. Massachusetts: Sinauer Associates. pp. 95, 147, 148. ISBN 9780878936953.
- "CALM1 – Calmodulin – Homo sapiens (Human) – CALM1 gene & protein". www.uniprot.org. Retrieved 2016-02-23.
- "Calmodulin". pdb101.rcsb.org. Retrieved 2016-02-23.
- PDB: 2HQW; Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea MA (December 2007). "The NMDA receptor NR1 C1 region bound to calmodulin: structural insights into functional differences between homologous domains". Structure (London, England : 1993). 15 (12): 1603–17. doi:10.1016/j.str.2007.10.012. PMC 2246159. PMID 18073110.
- PDB: 4QNH; Zhang M, Meng XY, Cui M, Pascal JM, Logothetis DE, Zhang JF (September 2014). "Selective phosphorylation modulates the PIP2 sensitivity of the CaM-SK channel complex". Nature Chemical Biology. 10 (9): 753–9. doi:10.1038/nchembio.1592. PMC 4420199. PMID 25108821.
- "Calmodulin". collab.itc.virginia.edu. Retrieved 2016-02-23.
- "Calmodulin Target Database". calcium.uhnres.utoronto.ca. Retrieved 2016-02-01.
- Chou JJ, Li S, Klee CB, Bax A (November 2001). "Solution structure of Ca(2+)-calmodulin reveals flexible hand-like properties of its domains". Nature Structural Biology. 8 (11): 990–7. doi:10.1038/nsb1101-990. PMID 11685248.
- Wang Q; et al. (2013). "Protein Recognition and Selection through Conformational and Mutually Induced Fit". Proceedings of the National Academy of Sciences. 110 (51): 20545–50. doi:10.1073/pnas.1312788110. PMC 3870683.
- "Home Page for Calmodulin". structbio.vanderbilt.edu. Retrieved 2016-02-23.
- McDowall, Jennifer. "Calmodulin". InterPro Protein Archive. Retrieved 23 February 2016.
- Tansey MG, Luby-Phelps K, Kamm KE, Stull JT (April 1994). "Ca(2+)-dependent phosphorylation of myosin light chain kinase decreases the Ca2+ sensitivity of light chain phosphorylation within smooth muscle cells". The Journal of Biological Chemistry. 269 (13): 9912–20. PMID 8144585.
- Walsh MP (June 1994). "Calmodulin and the regulation of smooth muscle contraction". Molecular and Cellular Biochemistry. 135 (1): 21–41. doi:10.1007/bf00925958. PMID 7816054.
- Martinsen A, Dessy C, Morel N (2014-10-31). "Regulation of calcium channels in smooth muscle: new insights into the role of myosin light chain kinase". Channels. 8 (5): 402–13. doi:10.4161/19336950.2014.950537. PMC 4594426. PMID 25483583.
- Nishizawa Y, Okui Y, Inaba M, Okuno S, Yukioka K, Miki T, et al. (October 1988). "Calcium/calmodulin-mediated action of calcitonin on lipid metabolism in rats". The Journal of Clinical Investigation. 82 (4): 1165–72. doi:10.1172/jci113713. PMC 442666. PMID 2844851.
- Lledo PM, Hjelmstad GO, Mukherji S, Soderling TR, Malenka RC, Nicoll RA (November 1995). "Calcium/calmodulin-dependent kinase II and long-term potentiation enhance synaptic transmission by the same mechanism". Proceedings of the National Academy of Sciences of the United States of America. 92 (24): 11175–9. Bibcode:1995PNAS...9211175L. doi:10.1073/pnas.92.24.11175. PMC 40594. PMID 7479960.
- Ranty B, Aldon D, Galaud JP (May 2006). "Plant calmodulins and calmodulin-related proteins: multifaceted relays to decode calcium signals". Plant Signaling & Behavior. 1 (3): 96–104. doi:10.4161/psb.1.3.2998. PMC 2635005. PMID 19521489.
- Chiasson D, Ekengren SK, Martin GB, Dobney SL, Snedden WA (August 2005). "Calmodulin-like proteins from Arabidopsis and tomato are involved in host defense against Pseudomonas syringae pv. tomato". Plant Molecular Biology. 58 (6): 887–897. doi:10.1007/s11103-005-8395-x. PMID 16240180.
- Leba LJ, Cheval C, Ortiz-Martín I, Ranty B, Beuzón CR, Galaud JP, Aldon D (September 2012). "CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway". The Plant Journal. 71 (6): 976–89. doi:10.1111/j.1365-313x.2012.05045.x. PMID 22563930.
- Sanchez AC, Subudhi PK, Rosenow DT, Nguyen HT (2002). "Mapping QTLs associated with drought resistance in sorghum (Sorghum bicolor L. Moench)". Plant Molecular Biology. 48 (5–6): 713–26. doi:10.1023/a:1014894130270. PMID 11999845.
- Singh S, Virdi AS, Jaswal R, Chawla M, Kapoor S, Mohapatra SB, et al. (June 2017). "A temperature-responsive gene in sorghum encodes a glycine-rich protein that interacts with calmodulin". Biochimie. 137 (Supplement C): 115–123. doi:10.1016/j.biochi.2017.03.010. PMID 28322928.
External links
- RCSB details...
- Nelson M, Chazin W. "Home Page for Calmodulin". EF-Hand Calcium-Binding Proteins Data Library. Vanderbilt University. Retrieved 2008-03-22.
- Ikura M. "Calmodulin Target Database". Ontario Cancer Institute, University of Toronto. PMID 12836676. Retrieved 2008-03-22. Cite journal requires
|journal=(help) - Calmodulin at the US National Library of Medicine Medical Subject Headings (MeSH)
- InterPro: IPR015754