Z-DNA
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern (instead of to the right, like the more common B-DNA form). Z-DNA is thought to be one of three biologically active double-helical structures along with A- and B-DNA.
History
Left-handed DNA was first discovered by Robert Wells and colleagues, during their studies of a repeating polymer of inosine–cytosine.[1] They observed a "reverse" circular dichroism spectrum for such DNAs, and interpreted this (correctly) to mean that the strands wrapped around one another in a left-handed fashion. The relationship between Z-DNA and the more familiar B-DNA was indicated by the work of Pohl and Jovin,[2] who showed that the ultraviolet circular dichroism of poly(dG-dC) was nearly inverted in 4 M sodium chloride solution. The suspicion that this was the result of a conversion from B-DNA to Z-DNA was confirmed by examining the Raman spectra of these solutions and the Z-DNA crystals.[3] Subsequently, a crystal structure of "Z-DNA" was published which turned out to be the first single-crystal X-ray structure of a DNA fragment (a self-complementary DNA hexamer d(CG)3). It was resolved as a left-handed double helix with two antiparallel chains that were held together by Watson–Crick base pairs (see X-ray crystallography). It was solved by Andrew Wang, Alexander Rich, and coworkers in 1979 at MIT.[4] The crystallisation of a B- to Z-DNA junction in 2005[5] provided a better understanding of the potential role Z-DNA plays in cells. Whenever a segment of Z-DNA forms, there must be B–Z junctions at its two ends, interfacing it to the B-form of DNA found in the rest of the genome.
In 2007, the RNA version of Z-DNA, Z-RNA, was described as a transformed version of an A-RNA double helix into a left-handed helix.[6] The transition from A-RNA to Z-RNA, however, was already described in 1984.[7]
Structure
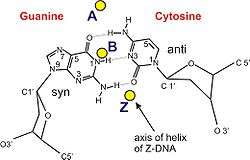
Z-DNA is quite different from the right-handed forms. In fact, Z-DNA is often compared against B-DNA in order to illustrate the major differences. The Z-DNA helix is left-handed and has a structure that repeats every other base pair. The major and minor grooves, unlike A- and B-DNA, show little difference in width. Formation of this structure is generally unfavourable, although certain conditions can promote it; such as alternating purine–pyrimidine sequence (especially poly(dGC)2), negative DNA supercoiling or high salt and some cations (all at physiological temperature, 37 °C, and pH 7.3–7.4). Z-DNA can form a junction with B-DNA (called a "B-to-Z junction box") in a structure which involves the extrusion of a base pair.[8] The Z-DNA conformation has been difficult to study because it does not exist as a stable feature of the double helix. Instead, it is a transient structure that is occasionally induced by biological activity and then quickly disappears.[9]
Predicting Z-DNA structure
It is possible to predict the likelihood of a DNA sequence forming a Z-DNA structure. An algorithm for predicting the propensity of DNA to flip from the B-form to the Z-form, ZHunt, was written by P. Shing Ho in 1984 at MIT.[10] This algorithm was later developed by Tracy Camp, P. Christoph Champ, Sandor Maurice, and Jeffrey M. Vargason for genome-wide mapping of Z-DNA (with Ho as the principal investigator).[11]
Biological significance
While no definitive biological significance of Z-DNA has been found, it is commonly believed to provide torsional strain relief during transcription, and it is associated with negative supercoiling.[5][12] However, while supercoiling is associated with both DNA transcription and replication, Z-DNA formation is primarily linked to the rate of transcription.[13]
A study of human chromosome 22 showed a correlation between Z-DNA forming regions and promoter regions for nuclear factor I. This suggests that transcription in some human genes may be regulated by Z-DNA formation and nuclear factor I activation.[11]
Z-DNA sequences downstream of promoter regions have been shown to stimulate transcription. The greatest increase in activity is observed when the Z-DNA sequence is placed three helical turns after the promoter sequence. Furthermore, Z-DNA is unlikely to form nucleosomes, which are often located after a Z-DNA forming sequence. Because of this property, Z-DNA is hypothesized to code for nucleosome positioning. Since the placement of nucleosomes influences the binding of transcription factors, Z-DNA is thought to regulate the rate of transcription.[14]
Developed behind the pathway of RNA polymerase through negative supercoiling, Z-DNA formed via active transcription has been shown to increase genetic instability, creating a propensity towards mutagenesis near promoters.[15] A study on Escherichia coli found that gene deletions spontaneously occur in plasmid regions containing Z-DNA-forming sequences.[16] In mammalian cells, the presence of such sequences was found to produce large genomic fragment deletions due to chromosomal double-strand breaks. Both of these genetic modifications have been linked to the gene translocations found in cancers such as leukemia and lymphoma, since breakage regions in tumor cells have been plotted around Z-DNA-forming sequences.[15] However, the smaller deletions in bacterial plasmids have been associated with replication slippage, while the larger deletions associated with mammalian cells are caused by non-homologous end-joining repair, which is known to be prone to error.[15][16]
The toxic effect of ethidium bromide (EtBr) on trypanosomas is caused by shift of their kinetoplastid DNA to Z-form. The shift is caused by intercalation of EtBr and subsequent loosening of DNA structure that leads to unwinding of DNA, shift to Z-form and inhibition of DNA replication.[17]
Z-DNA formed after transcription initiation
The first domain to bind Z-DNA with high affinity was discovered in ADAR1 using an approach developed by Alan Herbert.[18][19] Crystallographic and NMR studies confirmed the biochemical findings that this domain bound Z-DNA in a non-sequence-specific manner.[20][21][22] Related domains were identified in a number of other proteins through sequence homology.[19] The identification of the Zα domain provided a tool for other crystallographic studies that lead to the characterization of Z-RNA and the B–Z junction. Biological studies suggested that the Z-DNA binding domain of ADAR1 may localize this enzyme that modifies the sequence of the newly formed RNA to sites of active transcription.[23][24]
In 2003, Alex Rich noticed that a poxvirus virulence factor called E3L, that has a Zα related domain, mimicked a mammalian protein that binds Z-DNA.[25][26] In 2005, Rich and his colleagues pinned down what E3L does for the poxvirus. When expressed in human cells, E3L increases by five- to tenfold the production of several genes that block a cell's ability to self-destruct in response to infection. Rich speculates that the Z-DNA is necessary for transcription and that E3L stabilizes the Z-DNA, thus prolonging expression of the antiapoptotic genes. He suggests that a small molecule that interferes with the E3L binding to Z-DNA could thwart the activation of these genes and help protect people from pox infections.
Comparison geometries of some DNA forms
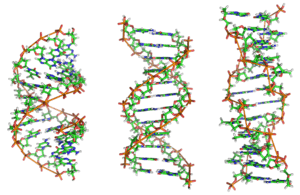
| A-form | B-form | Z-form | |
|---|---|---|---|
| Helix sense | right-handed | right-handed | left-handed |
| Repeating unit | 1 bp | 1 bp | 2 bp |
| Rotation/bp | 32.7° | 34.3° | 30° |
| bp/turn | 11 | 10 | 12 |
| Inclination of bp to axis | +19° | −1.2° | −9° |
| Rise/bp along axis | 2.3 Å (0.23 nm) | 3.32 Å (0.332 nm) | 3.8 Å (0.38 nm) |
| Pitch/turn of helix | 28.2 Å (2.82 nm) | 33.2 Å (3.32 nm) | 45.6 Å (4.56 nm) |
| Mean propeller twist | +18° | +16° | 0° |
| Glycosyl angle | anti | anti | C: anti, G: syn |
| Sugar pucker | C3′-endo | C2′-endo | C: C2′-endo, G: C3′-endo |
| Diameter | 23 Å (2.3 nm) | 20 Å (2.0 nm) | 18 Å (1.8 nm) |
See also
References
- ↑ Mitsui, Y.; Langridge, R.; Shortle, B. E.; Cantor, C. R.; Grant, R. C.; Kodama, M.; Wells, R. D. (1970). "Physical and enzymatic studies on poly d(I–C)·poly d(I–C), an unusual double-helical DNA". Nature. 228 (5277): 1166–1169. doi:10.1038/2281166a0. PMID 4321098.
- ↑ Pohl, F. M.; Jovin, T. M. (1972). "Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly(dG-dC)". Journal of Molecular Biology. 67 (3): 375–396. doi:10.1016/0022-2836(72)90457-3. PMID 5045303.
- ↑ Thamann, T. J.; Lord, R. C.; Wang, A. H.; Rich, A. (1981). "High salt form of poly(dG–dC)·poly(dG–dC) is left handed Z-DNA: raman spectra of crystals and solutions". Nucleic Acids Research. 9 (20): 5443–5457. doi:10.1093/nar/9.20.5443. PMC 327531. PMID 7301594.
- ↑ Wang, A. H.; Quigley, G. J.; Kolpak, F. J.; Crawford, J. L.; van Boom, J. H.; van der Marel, G.; Rich, A. (1979). "Molecular structure of a left-handed double helical DNA fragment at atomic resolution". Nature. 282 (5740): 680–686. Bibcode:1979Natur.282..680W. doi:10.1038/282680a0. PMID 514347.
- 1 2 Ha, S. C.; Lowenhaupt, K.; Rich, A.; Kim, Y. G.; Kim, K. K. (2005). "Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases". Nature. 437 (7062): 1183–1186. Bibcode:2005Natur.437.1183H. doi:10.1038/nature04088. PMID 16237447.
- ↑ Placido, D.; Brown, B. A., II; Lowenhaupt, K.; Rich, A.; Athanasiadis, A. (2007). "A left-handed RNA double helix bound by the Zalpha domain of the RNA-editing enzyme ADAR1". Structure. 15 (4): 395–404. doi:10.1016/j.str.2007.03.001. PMC 2082211. PMID 17437712.
- ↑ Hall, K.; Cruz, P.; Tinoco, I., Jr; Jovin, T. M.; van de Sande, J. H. (Oct 1984). "'Z-RNA'—a left-handed RNA double helix". Nature. 311 (5986): 584–586. Bibcode:1984Natur.311..584H. doi:10.1038/311584a0. PMID 6482970.
- ↑ de Rosa, M.; de Sanctis, D.; Rosario, A. L.; Archer, M.; Rich, A.; Athanasiadis, A.; Carrondo, M. A. (May 2010). "Crystal structure of a junction between two Z-DNA helices". Proceedings of the National Academy of Sciences. 107 (20): 9088–9092. Bibcode:2010PNAS..107.9088D. doi:10.1073/pnas.1003182107. PMC 2889044. PMID 20439751.
- ↑ Zhang, H.; Yu, H.; Ren, J.; Qu, X. (2006). "Reversible B/Z-DNA transition under the low salt condition and non-B-form poly(dA)poly(dT) selectivity by a cubane-like europium-L-aspartic acid complex". Biophysical Journal. 90 (9): 3203–3207. Bibcode:2006BpJ....90.3203Z. doi:10.1529/biophysj.105.078402. PMC 1432110. PMID 16473901.
- ↑ Ho, P. S.; Ellison, M. J.; Quigley, G. J.; Rich, A. (1986). "A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences". EMBO Journal. 5 (10): 2737–2744. PMC 1167176. PMID 3780676.
- 1 2 Champ, P. C.; Maurice, S.; Vargason, J. M.; Camp, T.; Ho, P. S. (2004). "Distributions of Z-DNA and nuclear factor I in human chromosome 22: a model for coupled transcriptional regulation". Nucleic Acids Research. 32 (22): 6501–6510. doi:10.1093/nar/gkh988. PMC 545456. PMID 15598822.
- ↑ Rich, A; Zhang, S (2003). "Timeline: Z-DNA: the long road to biological function". Nature Reviews Genetics. 4 (7): 566–572. doi:10.1038/nrg1115. PMID 12838348.
- ↑ Wittig, B.; Dorbic, T.; Rich, A. (1991). "Transcription is associated with Z-DNA formation in metabolically active permeabilized mammalian cell nuclei". Proceedings of the National Academy of Sciences. 88 (6): 2259–2263. Bibcode:1991PNAS...88.2259W. doi:10.1073/pnas.88.6.2259. PMC 51210. PMID 2006166.
- ↑ Wong, B.; Chen, S.; Kwon, J.-A.; Rich, A. (2007). "Characterization of Z-DNA as a nucleosome-boundary element in yeast Saccharomyces cerevisiae". Proceedings of the National Academy of Sciences. 104 (7): 2229–2234. Bibcode:2007PNAS..104.2229W. doi:10.1073/pnas.0611447104. PMC 1892989. PMID 17284586.
- 1 2 3 Wang, G.; Christensen, L. A.; Vasquez, K. M. (2006). "Z-DNA-forming sequences generate large-scale deletions in mammalian cells". Proceedings of the National Academy of Sciences. 108 (8): 2677–2682. Bibcode:2006PNAS..103.2677W. doi:10.1073/pnas.0511084103. PMC 1413824. PMID 16473937.
- 1 2 Freund, A. M.; Bichara, M.; Fuchs, R. P. (1989). "Z-DNA-forming sequences are spontaneous deletion hot spots". Proceedings of the National Academy of Sciences. 86 (19): 7465–7469. Bibcode:1989PNAS...86.7465F. doi:10.1073/pnas.86.19.7465. PMC 298085. PMID 2552445.
- ↑ Roy Chowdhury, A.; Bakshi, R.; Wang, J.,; Yıldırır, G.; Liu, B.; Pappas-Brown, V.; Tolun, G.; Griffith, J. D.; Shapiro, T. A.; Jensen, R. E.; Englund, P. T. (Dec 2010). "The killing of African trypanosomes by ethidium bromide". PLoS Pathogens. 6 (12): e1001226. doi:10.1371/journal.ppat.1001226. PMC 3002999. PMID 21187912.
- ↑ Herbert, A.; Rich, A. (1993). "A method to identify and characterize Z-DNA binding proteins using a linear oligodeoxynucleotide". Nucleic Acids Research. 21 (11): 2669–2672. doi:10.1093/nar/21.11.2669. PMC 309597. PMID 8332463.
- 1 2 Herbert, A.; Alfken, J.; Kim, Y. G.; Mian, I. S.; Nishikura, K.; Rich, A. (1997). "A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase". Proceedings of the National Academy of Sciences. 94 (16): 8421–8426. Bibcode:1997PNAS...94.8421H. doi:10.1073/pnas.94.16.8421. PMC 22942. PMID 9237992.
- ↑ Herbert, A.; Schade, M.; Lowenhaupt, K.; Alfken, J; Schwartz, T.; Shlyakhtenko, L. S.; Lyubchenko, Y. L.; Rich, A. (1998). "The Zα domain from human ADAR1 binds to the Z-DNA conformer of many different sequences". Nucleic Acids Research. 26 (15): 2669–2672. doi:10.1093/nar/26.15.3486. PMC 147729. PMID 9671809.
- ↑ Schwartz, T.; Rould, M. A.; Lowenhaupt, K.; Herbert, A.; Rich, A. (1999). "Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA". Science. 284 (5421): 1841–1845. doi:10.1126/science.284.5421.1841. PMID 10364558.
- ↑ Schade, M.; Turner, C. J.; Kühne, R.; Schmieder, P.; Lowenhaupt, K.; Herbert, A.; Rich, A.; Oschkinat, H (1999). "The solution structure of the Zα domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA". Proceedings of the National Academy of Sciences. 96 (22): 2465–2470. Bibcode:1999PNAS...9612465S. doi:10.1073/pnas.96.22.12465. PMC 22950. PMID 10535945.
- ↑ Herbert, A.; Rich, A. (2001). "The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1". Proceedings of the National Academy of Sciences. 98 (21): 12132–12137. Bibcode:2001PNAS...9812132H. doi:10.1073/pnas.211419898. PMC 59780. PMID 11593027.
- ↑ Halber, D. (1999-09-11). "Scientists observe biological activities of 'left-handed' DNA". MIT News Office. Retrieved 2008-09-29.
- ↑ Kim, Y. G.; Muralinath, M.; Brandt, T.; Pearcy, M.; Hauns, K.; Lowenhaupt, K.; Jacobs, B. L.; Rich, A. (2003). "A role for Z-DNA binding in vaccinia virus pathogenesis". Proceedings of the National Academy of Sciences. 100 (12): 6974–6979. Bibcode:2003PNAS..100.6974K. doi:10.1073/pnas.0431131100. PMC 165815. PMID 12777633.
- ↑ Kim, Y. G.; Lowenhaupt, K.; Oh, D. B.; Kim, K. K.; Rich, A. (2004). "Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: Implications for development of a therapy for poxvirus infection". Proceedings of the National Academy of Sciences. 101 (6): 1514–1518. Bibcode:2004PNAS..101.1514K. doi:10.1073/pnas.0308260100. PMC 341766. PMID 14757814.
- ↑ Sinden, Richard R. (1994). DNA Structure and Function (1st ed.). Academic Press. p. 398. ISBN 0-126-45750-6.
- ↑ Rich, A.; Norheim, A.; Wang, A. H. (1984). "The chemistry and biology of left-handed Z-DNA". Annual Review of Biochemistry. 53 (1): 791–846. doi:10.1146/annurev.bi.53.070184.004043. PMID 6383204.
- ↑ Ho, P. S. (1994-09-27). "The non-B-DNA structure of d(CA/TG)n does not differ from that of Z-DNA". Proceedings of the National Academy of Sciences. 91 (20): 9549–9553. Bibcode:1994PNAS...91.9549H. doi:10.1073/pnas.91.20.9549. PMC 44850. PMID 7937803.
