Legumain
| Legumain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
 | |||||||||
| Identifiers | |||||||||
| EC number | 3.4.22.34 | ||||||||
| CAS number | 149371-18-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Legumain (EC 3.4.22.34, asparaginyl endopeptidase, citvac, proteinase B, hemoglobinase, PRSC1 gene product or LGMN (Homo sapiens), vicilin peptidohydrolase, bean endopeptidase) is an enzyme that in humans is encoded by the LGMN gene (previous symbol PRSC1).[1][2][3]
Distribution
This enzyme was originally identified in the vacuoles of legume seeds, and was subsequently identified the lysosomes of mammals and Schistosoma mansoni.[4] They are now known to be present in a range of plants and animals.[5][6]
Activity
Reaction and specificity
This enzyme catalyses the following chemical reaction:
- Hydrolysis of proteins and small molecule substrates at -Asn-Xaa- bonds
Both plant and animal legumains are most active in acidic environments.[7][8]
Prodomain processing
Legmains are produced as inactive precursor zymogens. their C-terminal domain binds over their active site (where a substrate would normally bind), inhibiting activity.[7] Once in the acidic environment of the vacuole or lysosome, the prodomain is cleaved off to reveal the active enzyme.[7][8]
Mechanism
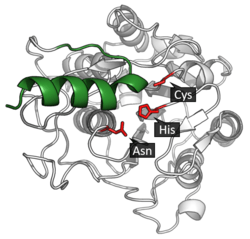
Legumain is a cysteine protease from the C13 family of the CD clan of proteases (MEROPS).[8] It uses a catalytic triad of Cysteine-Histidine-Asparagine in its active site to perform covalent proteolysis of its substrate.[3]

References
- ↑ Hara-Nishimura I (1998). "Asparaginyl endopeptidase". In Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. London: Academic Press. pp. 746–749.
- ↑ Dalton JP, Brindley PJ (1998). "Schistosome legumain". In Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. London: Academic Press. pp. 749–754.
- 1 2 Chen JM, Rawlings ND, Stevens RA, Barrett AJ (December 1998). "Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases". FEBS Letters. 441 (3): 361–5. doi:10.1016/S0014-5793(98)01574-9. PMID 9891971.
- ↑ Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, Stevens RA, Hewitt E, Watts C, Barrett AJ (March 1997). "Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase". The Journal of Biological Chemistry. 272 (12): 8090–8. doi:10.1074/jbc.272.12.8090. PMID 9065484.
- ↑ Müntz K, Shutov AD (August 2002). "Legumains and their functions in plants". Trends in Plant Science. 7 (8): 340–4. doi:10.1016/S1360-1385(02)02298-7. PMID 12167328.
- ↑ Shutov AD, Blattner FR, Kakhovskaya IA, Müntz K (February 2012). "New aspects of the molecular evolution of legumains, Asn-specific cysteine proteinases". Journal of Plant Physiology. 169 (3): 319–21. doi:10.1016/j.jplph.2011.11.005. PMID 22196948.
- 1 2 3 Chen JM, Fortunato M, Barrett AJ (December 2000). "Activation of human prolegumain by cleavage at a C-terminal asparagine residue". The Biochemical Journal. 352 Pt 2 (2): 327–34. doi:10.1042/bj3520327. PMC 1221463. PMID 11085925.
- 1 2 3 "C13 family". merops.sanger.ac.uk. MEROPS - the Peptidase Database. Retrieved 2016-06-09.
External links
- Legumain at the US National Library of Medicine Medical Subject Headings (MeSH)