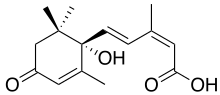
Abscisic acid
 | |
| Names | |
|---|---|
| Systematic IUPAC name
(2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid[1] | |
| Other names
(2Z,4E)-(S)-5-(1-Hydroxy-2,6,6-trimethyl-4-oxo-2-cyclohexen-1-yl)-3-methyl-2,4-pentanedienoic acid; Dormic acid; Dormin | |
| Identifiers | |
3D model (JSmol) |
|
| 3DMet | B00898 |
| Abbreviations | ABA |
| 2698956 | |
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.040.275 |
| EC Number | 244-319-5 |
| MeSH | Abscisic+Acid |
PubChem CID |
|
| RTECS number | RZ2475100 |
| |
| |
| Properties | |
| C15H20O4 | |
| Molar mass | 264.32 g·mol−1 |
| Appearance | Colorless crystals |
| Density | 1.193 g/mL |
| Melting point | 163 °C (325 °F; 436 K)[2] |
| log P | 1.896 |
| Acidity (pKa) | 4.868 |
| Basicity (pKb) | 9.129 |
| Hazards | |
| S-phrases (outdated) | S22, S24/25 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Abscisic acid (ABA) is a plant hormone. ABA functions in many plant developmental processes, including seed and bud dormancy, the control of organ size and stomatal closure. It is especially important for plants in the response to environmental stresses, including drought, soil salinity, cold tolerance, freezing tolerance, heat stress and heavy metal ion tolerance.[3]
In plants
Function
ABA was originally believed to be involved in abscission. This is now known to be the case only in a small number of plants. ABA-mediated signaling also plays an important part in plant responses to environmental stress and plant pathogens.[4][5] The plant genes for ABA biosynthesis and sequence of the pathway have been elucidated.[6][7] ABA is also produced by some plant pathogenic fungi via a biosynthetic route different from ABA biosynthesis in plants.[8]
Abscisic acid owes its names to its role in the abscission of plant leaves. In preparation for winter, ABA is produced in terminal buds. [9] This slows plant growth and directs leaf primordia to develop scales to protect the dormant buds during the cold season. ABA also inhibits the division of cells in the vascular cambium, adjusting to cold conditions in the winter by suspending primary and secondary growth.
Abscisic acid is also produced in the roots in response to decreased soil water potential (which is associated with dry soil) and other situations in which the plant may be under stress. ABA then translocates to the leaves, where it rapidly alters the osmotic potential of stomatal guard cells, causing them to shrink and stomata to close. The ABA-induced stomatal closure reduces transpiration (evaporation of water out of the stomata), thus preventing further water loss from the leaves in times of low water availability. A close linear correlation was found between the ABA content of the leaves and their conductance (stomatal resistance) on a leaf area basis.[10]
Seed germination is inhibited by ABA in antagonism with gibberellin. ABA also prevents loss of seed dormancy.
Several ABA-mutant Arabidopsis thaliana plants have been identified and are available from the Nottingham Arabidopsis Stock Centre - both those deficient in ABA production and those with altered sensitivity to its action. Plants that are hypersensitive or insensitive to ABA show phenotypes in seed dormancy, germination, stomatal regulation, and some mutants show stunted growth and brown/yellow leaves. These mutants reflect the importance of ABA in seed germination and early embryo development.
Pyrabactin (a pyridyl containing ABA activator) is a naphthalene sulfonamide hypocotyl cell expansion inhibitor, which is an agonist of the seed ABA signaling pathway.[11] It is the first agonist of the ABA pathway that is not structurally related to ABA.
Homeostasis
Biosynthesis
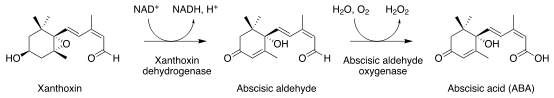
Abscisic acid (ABA) is an isoprenoid plant hormone, which is synthesized in the plastidal 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway; unlike the structurally related sesquiterpenes, which are formed from the mevalonic acid-derived precursor farnesyl diphosphate (FDP), the C15 backbone of ABA is formed after cleavage of C40 carotenoids in MEP. Zeaxanthin is the first committed ABA precursor; a series of enzyme-catalyzed epoxidations and isomerizations via violaxanthin, and final cleavage of the C40 carotenoid by a dioxygenation reaction yields the proximal ABA precursor, xanthoxin, which is then further oxidized to ABA. via abscisic aldehyde.[6]
Abamine has been designed, synthesized, developed and then patented as the first specific ABA biosynthesis inhibitor, which makes it possible to regulate endogenous levels of ABA.[12]
Location and timing of ABA biosynthesis
- Released during desiccation of the vegetative tissues and when roots encounter soil compaction.[13]
- Synthesized in green fruits at the beginning of the winter period
- Synthesized in maturing seeds, establishing dormancy
- Mobile within the leaf and can be rapidly translocated from the roots to the leaves by the transpiration stream in the xylem
- Produced in response to environmental stress, such as heat stress, water stress, salt stress
- Synthesized in all plant parts, e.g., roots, flowers, leaves and stems
Inactivation
ABA can be catabolized to phaseic acid via CYP707A (a group of P450 enzymes) or inactivated by glucose conjugation (ABA-glucose ester) via the enzyme AOG. Catabolism via the CYP707As is very important for ABA homeostasis, and mutants in those genes generally accumulate higher levels of ABA than lines overexpressing ABA biosynthetic genes.[14] In soil bacteria, an alternative catabolic pathway leading to dehydrovomifoliol via the enzyme vomifoliol dehydrogenase has been reported.
Effects
- Antitranspirant - Induces stomatal closure, decreasing transpiration to prevent water loss.[15]
- Inhibits fruit ripening
- Responsible for seed dormancy by inhibiting cell growth – inhibits seed germination
- Inhibits the synthesis of Kinetin nucleotide[16]
- Downregulates enzymes needed for photosynthesis.[17]
- Acts on endodermis to prevent growth of roots when exposed to salty conditions[18]
- Delays cell division
Signal cascade

In the absence of ABA, the phosphatase ABI1-INSENSITIVE1 (ABI1) inhibits the action of SNF1-related protein kinases (subfamily 2) (SnRK2s). ABA is perceived by the PYRABACTIN RESISTANCE 1 (PYR1) and PYR1-like membrane proteins. On ABA binding, PYR1 binds to and inhibits ABI1. When SnRK2s are released from inhibition, they activate several transcription factors from the ABA RESPONSIVE ELEMENT-BINDING FACTOR (ABF) family. ABFs then go on to cause changes in the expression of a large number of genes. [3] Around 10% of plant genes are thought to be regulated by ABA. [19]
In fungi
Like plants, some fungal species (for example Cercospora rosicola, Botrytis cinerea [20] and Magnaporthe oryzae) have an endogenous biosynthesis pathway for ABA. In fungi, it seems to be the MVA biosynthetic pathway that is predominant (rather than the MEP pathway that is responsible for ABA biosynthesis in plants). One role of ABA produced by these pathogens seems to be to suppress the plant immune responses.
In animals
ABA has also been found to be present in metazoans, from sponges up to mammals including humans.[21] Currently, its biosynthesis and biological role in animals is poorly known. ABA has recently been shown to elicit potent anti-inflammatory and anti-diabetic effects in mouse models of diabetes/obesity, inflammatory bowel disease, atherosclerosis and influenza infection.[22] Many biological effects in animals have been studied using ABA as a nutraceutical or pharmacognostic drug, but ABA is also generated endogenously by some cells (like macrophages) when stimulated. There are also conflicting conclusions from different studies, where some claim that ABA is essential for pro-inflammatory responses whereas other show anti-inflammatory effects. Like with many natural substances with medical properties, ABA has become popular also in naturopathy. While ABA clearly has beneficial biological activities and many naturopathic remedies will contain high levels of ABA (such as wheatgrass juice, fruits and vegetables), some of the health claims made may be exaggerated or overly optimistic. In mammalian cells ABA targets a protein known as lanthionine synthetase C-like 2 (LANCL2), triggering an alternative mechanism of activation of peroxisome proliferator-activated receptor gamma (PPAR gamma).[23] LANCL2 is conserved in plants and was originally suggested to be an ABA receptor also in plants, which was later challenged.[24]
Measurement of ABA concentration
Several methods can help to quantify the concentration of abscisic acid in a variety of plant tissue. The quantitative methods used are based on HPLC and GC, and ELISA. Recently, 2 independent FRET probes have been developed that can measure intracellular ABA concentrations in real time in vivo.[25][26]
References
- ↑ "Abscisic Acid - Compound Summary". PubChem Compound. USA: National Center for Biotechnology Information. 16 September 2004. Identification and Related Records. Retrieved 22 October 2011.
- ↑ "ChemSpider database - Abscisic acid - Properties". Retrieved 27 December 2012. The melting point is decided by experimental data by Tokyo Chemical Industry Ltd.
- 1 2 Finkelstein, Ruth (2013-11-01). "Abscisic Acid Synthesis and Response". The Arabidopsis Book / American Society of Plant Biologists. 11. doi:10.1199/tab.0166. PMC 3833200. PMID 24273463.
- ↑ Zhu, Jian-Kang (2002). "Salt and Drought Stress Signal Transduction in Plants". Annual Review of Plant Biology. 53: 247–73. doi:10.1146/annurev.arplant.53.091401.143329. PMC 3128348. PMID 12221975.
- ↑ Seo, M; Koshiba, T (2002). "Complex regulation of ABA biosynthesis in plants". Trends in Plant Science. 7 (1): 41–8. doi:10.1016/S1360-1385(01)02187-2. PMID 11804826.
- 1 2 Nambara, Eiji; Marion-Poll, Annie (2005). "Abscisic Acid Biosynthesis and Catabolism". Annual Review of Plant Biology. 56: 165–85. doi:10.1146/annurev.arplant.56.032604.144046. PMID 15862093.
- ↑ Milborrow, B.V. (2001). "The pathway of biosynthesis of abscisic acid in vascular plants: A review of the present state of knowledge of ABA biosynthesis". Journal of Experimental Botany. 52 (359): 1145–64. doi:10.1093/jexbot/52.359.1145. PMID 11432933.
- ↑ Siewers, V.; Smedsgaard, J.; Tudzynski, P. (2004). "The P450 Monooxygenase BcABA1 is Essential for Abscisic Acid Biosynthesis in Botrytis cinerea". Applied and Environmental Microbiology. 70 (7): 3868–76. doi:10.1128/AEM.70.7.3868-3876.2004. PMC 444755. PMID 15240257.
- ↑ Wang, Dongling; Gao, Zhenzhen; Du, Peiyong; Xiao, Wei; Tan, Qiuping; Chen, Xiude; Li, Ling; Gao, Dongsheng (2016). "Expression of ABA Metabolism-Related Genes Suggests Similarities and Differences Between Seed Dormancy and Bud Dormancy of Peach (Prunus persica)". Frontiers in Plant Science. 6. doi:10.3389/fpls.2015.01248. ISSN 1664-462X.
- ↑ Steuer, Barbara; Thomas Stuhlfauth; Heinrich P. Fock (1988). "The efficiency of water use in water stressed plants is increased due to ABA induced stomatal closure". Photosynthesis Research. 18 (3): 327–336. doi:10.1007/BF00034837. ISSN 0166-8595. Retrieved 2012-08-10.
- ↑ Park, Sang-Youl; P. Fung; N. Nishimura; D. R. Jensen; H. Fuiji; Y. Zhao, S. Lumba; et al. (May 2009). "Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins". Science Signaling. 324 (5930). doi:10.1126/science.1173041. PMC 2827199.
- ↑ Abscisic acid biosynthesis inhibitor, Shigeo Yoshida et al US 7098365
- ↑ DeJong-Hughes, J., et al. (2001) Soil Compaction: causes, effects and control. University of Minnesota extension service
- ↑ Finkelstein, Ruth (November 2013). "Abscisic Acid Synthesis and Response". The Arabidopsis Book. 11: e0166. doi:10.1199/tab.0166. PMC 3833200. PMID 24273463.
- ↑ Zhang, Jianhua; Schurr, U.; Davies, W. J. (1987). "Control of Stomatal Behaviour by Abscisic Acid which Apparently Originates in the Roots". Journal of Experimental Botany. 38 (7): 1174–1181. doi:10.1093/jxb/38.7.1174.
- ↑ Miernyk, J. A. (1979). "Abscisic Acid Inhibition of Kinetin Nucleotide Formation in Germinating Lettuce Seeds". Physiologia Plantarum. 45: 63–6. doi:10.1111/j.1399-3054.1979.tb01664.x.
- ↑ Chandler, P M; Robertson, M (1994). "Gene Expression Regulated by Abscisic Acid and its Relation to Stress Tolerance". Annual Review of Plant Physiology and Plant Molecular Biology. 45: 113–41. doi:10.1146/annurev.pp.45.060194.000553.
- ↑ Duan, Lina; D. Dietrich; C. H. Ng; P. M. Y. Chan; R. Bhalerao; M. J. Bennett; J. R. Dinneny. (Jan 2013). "Endodermal ABA Signaling Promotes Lateral Root Quiescence during Salt Stress in Arabidopsis Seedlings" (PDF). The Plant Cell. 25 (1): 324–341. doi:10.1105/tpc.112.107227. PMC 3584545. PMID 23341337. Retrieved 11 January 2015.
- ↑ Fernando, V.C. Dilukshi; Schroeder, Dana F. (2016-02-17). Abiotic and Biotic Stress in Plants - Recent Advances and Future Perspectives. InTech. doi:10.5772/61957. ISBN 9789535122500.
- ↑ Sievers, Verena; Kokkelink, Leonie; Smedsgaard, Jørn; Tudzynski, Paul (July 2006). "Identification of an Abscisic Acid Gene Cluster in the Grey Mold Botrytis cinerea". Appl Environ Microbiol. 72: 4619–4626. doi:10.1128/AEM.02919-05. PMC 1489360. PMID 16820452.
- ↑ Na-Hang, Li; Rui-Lin, Hao; Shan-Shan, Wu; Peng-Cheng, Guo; Can-Jiang, Chen; Li-Ping, Pan; He, Ni (2011). "Occurrence, function and potential medicinal applications of the phytohormone abscisic acid in animals and humans". Biochemical Pharmacology. 82 (7): 701–712. doi:10.1016/j.bcp.2011.06.042. PMID 21763293.
- ↑ Bassaganya-Riera, J; Skoneczka, J; Kingston, DG; Krishnan, A; Misyak, SA; Guri, AJ; Pereira, A; Carter, AB; Minorsky, P; Tumarkin, R; Hontecillas, R (2010). "Mechanisms of action and medicinal applications of abscisic Acid". Current Medicinal Chemistry. 17 (5): 467–78. doi:10.2174/092986710790226110. PMID 20015036. Archived from the original on 2012-04-01. Retrieved 2018-09-30.
- ↑ Bassaganya-Riera, J.; Guri, A. J.; Lu, P.; Climent, M.; Carbo, A.; Sobral, B. W.; Horne, W. T.; Lewis, S. N.; Bevan, D. R.; Hontecillas, R. (2010). "Abscisic Acid Regulates Inflammation via Ligand-binding Domain-independent Activation of Peroxisome Proliferator-activated Receptor". Journal of Biological Chemistry. 286 (4): 2504–16. doi:10.1074/jbc.M110.160077. PMC 3024745. PMID 21088297.
- ↑ Chen, JG; Ellis, BE (2008). "GCR2 is a new member of the eukaryotic lanthionine synthetase component C-like protein family". Plant Signal Behav. 3: 307–10. doi:10.4161/psb.3.5.5292. PMC 2634266. PMID 19841654.
- ↑ Waadt, R; Hitomi, K; Nishimura, N; Hitomi, C; Adams, SR; Getzoff, ED; Schroeder, JI (2014). "FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis". eLife. 3: e01739. doi:10.7554/eLife.01739. PMC 3985518. PMID 24737861.
- ↑ Jones, AM; Danielson, JA; Manjokumar, SN; Laquar, V; Grossmann, G; Frommer, WB (2014). "Abscisic acid dynamics in roots detected with genetically encoded FRET sensors". eLife. 3: e01741. doi:10.7554/eLife.01741. PMC 3985517. PMID 24737862.