Paraspeckle
In cell biology, a Paraspeckle is an irregularly shaped compartment of the cell, approximately 0.2-1 μm in size,[1] found in the nucleus' interchromatin space.[2] First documented in HeLa cells, where there are generally 10-30 per nucleus,[3] Paraspeckles are now known to also exist in all human primary cells, transformed cell lines and tissue sections.[4] Their name is derived from their distribution in the nucleus; the "para" is short for parallel and the "speckle" refers to the splicing speckles to which they are always in close proximity.[3] Their function is still not fully understood, but they are thought to regulate gene expression by sequestrating proteins or mRNAs with inverted repeats in their 3′ UTRs.[5][6]
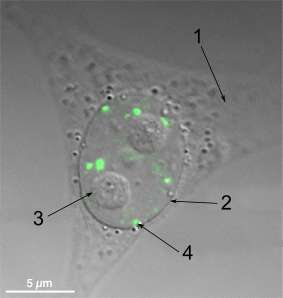
Structure
Paraspeckles are organised into core-shell spheroidal structures; seven proteins on a scaffold of lncRNA Neat1 (the 23kb isoform termed NEAT1_2 or NEAT1v2). [9] In 2016, West et al. proposed the currently accepted model for Paraspeckles. This was based on their current findings using super-resolution microscopy.[10] Their models state that the Neat1_2 scaffold folds into a V-shaped unit. Many of these units then are assembled into a core-shell spheroid by FUS proteins. Core proteins SFPQ, NONO and PSPC1 tightly associate to the assembled structure. Finally, the shell forms, composed of partially co-localised TDP43 proteins. Due to the integral nature of Neat1 to paraspeckles assembly, assembly is thought to occur in close proximity to Neat1 transcription sites.[9]
It has been noted that Paraspeckles have a great deal of commonality both in features and structures with cytoplasmic stress granules, another type of membrane-less organelle. This conclusion arose from the fact that both contain common component proteins[11], become more abundant with stress, seem to function through sequestering other proteins and both have distinct core or shell regions with predictable localised molecules[11].
Localization
Paraspeckles are dynamic structures that are altered in response to changes in cellular metabolic activity. They are transcription-dependent.[2] All five of the proposed protein components have RNA recognition motifs (RRMs)[3] and, in the absence of RNA polymerase II transcription, the Paraspeckle disappears and all of its associated proponents form a crescent shaped perinucleolar cap in the nucleolus. This phenomenon is demonstrated during the cell cycle. In the cell cycle, Paraspeckles are present during interphase and during all of mitosis except for telophase because, when the two daughter nuclei are formed, there is no RNA Pol II transcription so the protein components instead form a perinucleolar cap. The localization patterns were also duplicated in experiments using transcription inhibiting drugs.[4]
Function
The function of the paraspeckle nuclear domain, as a whole, is still not well understood. It has been postulated that the activity of p54nrb, a protein component, is dependent on its localization.[4] It is therefore possible that the paraspeckle's role is to provide ordered localization of its component proteins and to thereby help direct their activity. It has also been suggested that the paraspeckle contributes to transcriptional regulation.[12] Neither of these hypotheses, however, is universally accepted and therefore insight into the paraspeckle's larger role must be derived from the function of its protein components (PSP1, p54nrb, PSP2 and possibly CFI(m)68 and PSF).
The function of PSP1, the protein whose localization pattern led to the discovery of the paraspeckle,[2] is not well understood. Myojin et al. speculated that PSP1, which is highly concentrated in the testis, may regulate the germ cells' early mRNA processing and assist in chromatin remodeling and nuclear shaping during spermatogenesis.[13] PSP1 also forms a dimer with the second protein component: p54nrb. P54nrb has reported involvement in numerous nuclear events including "transcriptional regulation, splicing, DNA unwinding, nuclear retention of hyperedited dsRNA, viral RNA processing, control and cell proliferation, and circadian rhythm maintenance".[4] The final confirmed component, PSP2, is involved in RNA splicing and coactivates hormone receptors.[3]
Later studies have led to the identification of two additional proteins that are likely components of the paraspeckle. In 2004 Dettwiler et al. revealed CFI(m)68 as a possible component of the paraspeckle.[14] CFI(m)68 has been implicated with the preliminary step in pre-mRNA 3' end splicing. Fox et al.'s 2005 article also contains evidence of a possible fifth protein component of the paraspeckle: PSF.[4] PSF can bind both RNA and DNA and interacts with pre-mRNA splicing proteins that work in conjunction with proteins like CFI(m)68.[13] It can dimerize with p54nrb. Furthermore, it colocalizes with PSP1 both in the paraspeckle and, if in the presence of transcription inhibiting drugs, in the same perinucleolar cap.[4] If PSF is in fact part of the paraspeckle that would help further substantiate an assertion by Myojin, et al. that paraspeckle components may participate in pre-mRNA splicing.
In 2005 a new role for paraspeckles in a novel method for controlling gene expression was reported by Prasanth et al.[15] In this study, a nuclear enriched non-coding RNA (termed CTNRNA) was identified that specifically localised to paraspeckles in the nuclei of several cell types. The group found that the RNA was retained in the nucleus at paraspeckles, and was associated with the paraspeckle proteins P54nrb and PSP1, likely through direct interactions between the proteins and motifs in the 3' untranslated (3' UTR) region of the RNA. The CTN non-coding RNA is a longer transcript produced from a gene that also encodes the membrane protein MCAT2, a cationic amino acid transporter. When cells became stressed, the nuclear non-coding RNA levels were reduced, coupled with an increase in cytoplasmic signal for the MCAT2 mRNA and protein. This led the authors to speculate that the paraspeckles were effectively a storage site for the spliced and processed CTN RNA, that were able to release the RNA in a functional protein-encoding form when the cell received a signal. This shorter form was then free to be transported to the cytoplasm and used as a template for protein production. This 'Rapid Release Nuclear Retention mechanism' is thought to save the cell 25 minutes in the production of the mCAT2 protein, as the RNA has already been transcribed, processed and spliced whilst it is being held in the paraspeckles.
While paraspeckles are originally defined as nuclear bodies containing paraspeckle proteins, the field is re-defining these nuclear bodies as structures containing the long noncoding RNA Neat1_2.[16] Neat1_2 is expressed in a particular cell types in mouse tissues, and thus paraspeckles are not ubiquitous nuclear bodies in living animals.[17].
Paraspeckle Composition
| Gene Name | Importance in paraspeckle formation | Prionlike domain(a) | ALS Mutation(b) | Liquid-Liquid phase separation link(c) | Paraspeckle Zone(d) |
| PARASPECKLE PROTEINS | |||||
|---|---|---|---|---|---|
| AHDC1 | Dispensable | No | |||
| AKAP8L | Dispensable | Yes | |||
| CELF6 | n.d.(e) | No | |||
| CIRBP | Dispensable | No | |||
| CPSF5 | Dispensable | No | |||
| CPSF6 | Dispensable | No | |||
| CPSF7 | Important | No | |||
| DAZAP1 | Essential | Yes | |||
| DLX3 | n.d. | Yes | |||
| EWSR1 | Dispensable | Yes | Yes | ||
| FAM98A | Important | Yes | |||
| FIGN | Important | Yes | |||
| FUS | Essential | Yes | Yes | Yes | Core |
| HNRNPA1 | Important | Yes | Yes | Yes | |
| HNRNPA1L2 | n.d. | Yes | |||
| HNRNPF | n.d. | No | |||
| HNRNPH1 | n.d. | Yes | |||
| HNRNPH3 | Essential | Yes | |||
| HNRNPK | Essential | No | |||
| HNRNPR | Important | Yes | |||
| HNRNPUL1 | Important | Yes | |||
| MEX3A | n.d. | No | |||
| NONO | Essential | Yes | Core | ||
| PCED1A | Important | No | |||
| PSPC1 | Dispensable | Yes | Core | ||
| RBM3 | Dispensable | Yes | |||
| RBM4B | Dispensable | No | |||
| RBM7 | Dispensable | No | |||
| RBM12 | Important | Yes | |||
| RBM14 | Essential | Yes | Yes | Patch | |
| RBMX | Dispensable | No | |||
| RUNX3 | Dispensable | Yes | |||
| SFPQ | Essential | Yes | Yes | Core | |
| SMARCA4 (BRG1) | Essential | No | Patch | ||
| SRSF10 | Important | No | |||
| SS18L1 | n.d. | Yes | Yes | ||
| TAF14 | Important | Yes | Yes | ||
| TDP43 | n.d. | Yes | Yes | Shell | |
| UBAP2L | Dispensable | Yes | |||
| ZC3HG | Dispensable | Yes | |||
| PARASPECKLE RNAs | |||||
| NEAT1 | Essential | N/A(e) | 5' + 3' Shell, middle core | ||
| IR-containing RNAs | Dispensable | N/A | |||
| AG-rich RNAs | Dispensable | N/A | Shell | ||
| (a) A type of low complexity domain rich in polar and small amino acids (Gly, Ala, Ser, Pro, Asn, Gln, Tyr) implicated in forming fibrillar higher-order aggregates
(b) Amyotrophic lateral sclerosis, also known as motor neuron disease (c) Partition of components of molecular mixtures into distinct demixed phases (e.g., oil and water). In the cell, many membrane-less organelle display liquid behaviours suggesting that they are demixed liquids (d) The paraspeckle zones relate to the super-resolution imaging of paraspeckles (e) Abbreviations: n.d., not determined; N/A, not applicable | |||||
References
- Fox A (2007-03-07). "Paraspeckle Size" (Interview). Interviewed by Sundby R. E-mail Correspondence.
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI (January 2002). "Paraspeckles: a novel nuclear domain". Current Biology. 12 (1): 13–25. doi:10.1016/S0960-9822(01)00632-7. PMID 11790299. Archived from the original on 2012-12-08.
- Fox A, Bickmore W (2004). "Nuclear Compartments: Paraspeckles". Archived from the original on 2 May 2006. Retrieved 6 March 2007.
- Fox AH, Bond CS, Lamond AI (November 2005). "P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner". Molecular Biology of the Cell. 16 (11): 5304–15. doi:10.1091/mbc.E05-06-0587. PMC 1266428. PMID 16148043.
- Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. (February 2014). "Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli". Molecular Cell. 53 (3): 393–406. doi:10.1016/j.molcel.2014.01.009. PMID 24507715.
- Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, et al. (January 2014). "NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies". Molecular Biology of the Cell. 25 (1): 169–83. doi:10.1091/mbc.e13-09-0558. PMC 3873887. PMID 24173718.
- West JA, Mito M, Kurosaka S, Takumi T, Tanegashima C, Chujo T, et al. (September 2016). "Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization". The Journal of Cell Biology. 214 (7): 817–30. doi:10.1083/jcb.201601071. PMID 27646274.
- Hu SB, Yao RW, Chen LL (September 2016). "Shedding light on paraspeckle structure by super-resolution microscopy". The Journal of Cell Biology. 214 (7): 789–91. doi:10.1083/jcb.201609008. PMC 5037413. PMID 27646270.
- Hu SB, Yao RW, Chen LL (September 2016). "Shedding light on paraspeckle structure by super-resolution microscopy". The Journal of Cell Biology. 214 (7): 789–91. doi:10.1083/jcb.201609008. PMC 5037413. PMID 27646270.
- West JA, Mito M, Kurosaka S, Takumi T, Tanegashima C, Chujo T, et al. (September 2016). "Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization". The Journal of Cell Biology. 214 (7): 817–30. doi:10.1083/jcb.201601071. PMID 27646274.
- Fox, Archa H.; Nakagawa, Shinichi; Hirose, Tetsuro; Bond, Charles S. (February 2018). "Paraspeckles: Where Long Noncoding RNA Meets Phase Separation". Trends in Biochemical Sciences. 43 (2): 124–135. doi:10.1016/j.tibs.2017.12.001. ISSN 0968-0004. PMID 29289458.
- Schuldt A (February 2002). "Proteomics of the nucleolus". Nature Cell Biology. 4 (2): E35. doi:10.1038/ncb0202-e35. PMID 11835055.
- Myojin R, Kuwahara S, Yasaki T, Matsunaga T, Sakurai T, Kimura M, et al. (September 2004). "Expression and functional significance of mouse paraspeckle protein 1 on spermatogenesis". Biology of Reproduction. 71 (3): 926–32. doi:10.1095/biolreprod.104.028159. PMID 15140795.
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM (August 2004). "Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization" (PDF). The Journal of Biological Chemistry. 279 (34): 35788–97. doi:10.1074/jbc.M403927200. PMID 15169763.
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, et al. (October 2005). "Regulating gene expression through RNA nuclear retention". Cell. 123 (2): 249–63. doi:10.1016/j.cell.2005.08.033. PMID 16239143.
- Fox AH, Nakagawa S, Hirose T, Bond CS (February 2018). "Paraspeckles: Where Long Noncoding RNA Meets Phase Separation". Trends in Biochemical Sciences. 43 (2): 124–135. doi:10.1016/j.tibs.2017.12.001. PMID 29289458.
- Nakagawa S, Naganuma T, Shioi G, Hirose T (April 2011). "Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice". The Journal of Cell Biology. 193 (1): 31–9. doi:10.1083/jcb.201011110. PMID 21444682.
- Fox, Archa H.; Nakagawa, Shinichi; Hirose, Tetsuro; Bond, Charles S. (February 2018). "Paraspeckles: Where Long Noncoding RNA Meets Phase Separation". Trends in Biochemical Sciences. 43 (2): 124–135. doi:10.1016/j.tibs.2017.12.001. ISSN 0968-0004. PMID 29289458.
External links
- The Nuclear Compartments:Paraspeckle page on the Nuclear Protein Database, written by Dr. Archa Fox and Dr. Wendy Bickmore, provides a factsheet and links to information on paraspeckle components.
