Protein O-GlcNAcase
Protein O-GlcNAcase (EC 3.2.1.169, OGA, glycoside hydrolase O-GlcNAcase, O-GlcNAcase, BtGH84, O-GlcNAc hydrolase) is an enzyme with systematic name (protein)-3-O-(N-acetyl-D-glucosaminyl)-L-serine/threonine N-acetylglucosaminyl hydrolase.[5][6][7][8][9] OGA is encoded by the MGEA5 gene. This enzyme catalyses the removal of the O-GlcNAc post-translational modification in the following chemical reaction:
- [protein]-3-O-(N-acetyl-β-D-glucosaminyl)-L-serine + H2O ⇌ [protein]-L-serine + N-acetyl-D-glucosamine
- [protein]-3-O-(N-acetyl-β-D-glucosaminyl)-L-threonine + H2O ⇌ [protein]-L-threonine + N-acetyl-D-glucosamine
| OGA | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||
| Aliases | OGA, MEA5, NCOAT, meningioma expressed antigen 5 (hyaluronidase), MGEA5, O-GlcNAcase | ||||||||||||||||||||||||
| External IDs | OMIM: 604039 MGI: 1932139 HomoloGene: 8154 GeneCards: OGA | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | Chr 10: 101.78 – 101.82 Mb | Chr 19: 45.75 – 45.78 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Nomenclature
| Protein O-GlcNAcase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 3.2.1.169 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Other names include:
- Nuclear cytoplasmic O-GlcNAcase and acetyltransferase
Isoforms
There are three isoforms of O-GlcNAcase in humans that have been identified. Full-length O-GlcNAcase (fOGA), the shortest O-GlcNAcase (sOGA), and a variant of OGA (vOGA). The human OGA gene is capable of producing two separate transcriptions, each capable of encoding a different OGA isoform. The long isoform gene codes for fOGA, a bifunctional enzyme that primarily resides in the cytoplasm. In contrast, vOGA resides within the nucleus. However, all three isoforms exhibit glycoside hydrolase activity.[10]
Homologs
Protein O-GlcNAcases belong to glycoside hydrolase family 84 of the carbohydrate active enzyme classification.[11] Homologs exist in other species as O-GlcNAcase is conserved in higher eukaryotic species. In a pairwise alignment, humans share 55% homology with Drosophilia and 43% with C. elegans. Drosophilia and C. elegans share 43% homology. Among mammals, the OGA sequence is even more highly conserved. The mouse and the human have 97.8% homology. However, OGA does not share significant homology with other proteins. However, short stretches of about 200 amino acids in OGA have homology with some proteins such as hyaluronidase, a putative acetyltransferase, eukaryotic translation elongation factor-1γ, and the 11-1 polypeptide.[12]
Reaction
Protein O-GlcNAcylation
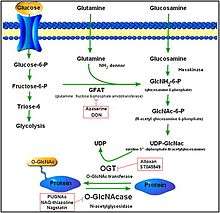
O-GlcNAcylation is a form of glycosylation, the site-specific enzymatic addition of saccharides to proteins and lipids. This form of glycosylation is with O-linked β-N-acetylglucosamine or β-O-linked 2-acetamido-2-deoxy-D-glycopyranose (O-GlcNAc). In this form, a single sugar (β-N-acetylglucosamine) is added to serine and threonine residues of nuclear or cytoplasmic proteins. Two conserved enzymes control this glycosylation of serine and threonine: O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA). While OGT catalyzes the addition of O-GlcNAc to serine and threonine, OGA catalyzes the hydrolytic cleavage of O-GlcNAc from post-transitionally modified proteins.[13]
OGA is a member of the family of hexosaminidases. However, unlike lysosomal hexosaminidases, OGA activity is the highest at neutral pH (approximately 7) and it localizes mainly to the cytosol. OGA and OGT are synthesized from two conserved genes (OGA is encoded by MGEA5) and are expressed throughout the human body with high levels in the brain and pancreas. The products of O-GlcNAc and the process itself plays a role in embryonic development, brain activity, hormone production, and a myriad of other activities.[14][15]
Over 600 proteins are targets for O-GlcNAcylation. While the functional effects of O-GlcNAc modification is not fully known, it is known that O-GlcNAc modification impacts many cellular activities such as lipid/carbohydrate metabolism and hexosamine biosynthesis. Modified proteins may modulate various downstream signaling pathways by influencing transcription and proteomic activities.[16]
Mechanism and Inhibition
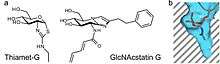
OGA catalyzes O-GlcNAc hydrolysis via an oxazoline reaction intermediate.[17] Stable compounds which mimic the reaction intermediate can act as selective enzyme inhibitors. Thiazoline derivatives of GlcNAc can be used as a reaction intermediate. An example of this includes Thiamet-G as shown on the right. A second form of inhibition can occur from the mimicry of the transition state. The GlcNAcstatin family of inhibitors exploit this mechanism in order to inhibit OGA activity. For both types of inhibitors, OGA can be selected apart from the generic lysosomal hexosaminidases by elongating the C2 substituent in their chemical structure. This takes advantage of a deep pocket in OGA's active site that allow it to bind analogs of GlcNAc.[18]
There is potential for regulation of O-GlcNAcase for the treatment of Alzheimer's disease. When the tau protein in the brain is hyperphosphorylated, neurofibrillary tangles form, which are a pathological hallmark for neurodegenerative diseases such as Alzheimer's disease. In order to treat this condition, OGA is targeted by inhibitors such as Thiamet-G in order to prevent O-GlcNAc from being removed from tau, which assists in preventing tau from becoming phosphorylated.[19]
Structure
X-ray structures are available for a range of O-GlcNAcase proteins. The X-ray structure of human O-GlcNAcase in complex with Thiamet-G identified the structural basis of enzyme inhibition.[20]
References
- GRCh38: Ensembl release 89: ENSG00000198408 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000025220 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Wells L, Gao Y, Mahoney JA, Vosseller K, Chen C, Rosen A, Hart GW (January 2002). "Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase". The Journal of Biological Chemistry. 277 (3): 1755–61. doi:10.1074/jbc.M109656200. PMID 11788610.
- Cetinbaş N, Macauley MS, Stubbs KA, Drapala R, Vocadlo DJ (March 2006). "Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants". Biochemistry. 45 (11): 3835–44. doi:10.1021/bi052370b. PMID 16533067.
- Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, et al. (April 2006). "Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity". Nature Structural & Molecular Biology. 13 (4): 365–71. doi:10.1038/nsmb1079. PMID 16565725.
- Kim EJ, Kang DO, Love DC, Hanover JA (June 2006). "Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate". Carbohydrate Research. 341 (8): 971–82. doi:10.1016/j.carres.2006.03.004. PMID 16584714.
- Dong DL, Hart GW (July 1994). "Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol". The Journal of Biological Chemistry. 269 (30): 19321–30. PMID 8034696.
- Li J, Huang CL, Zhang LW, Lin L, Li ZH, Zhang FW, Wang P (July 2010). "Isoforms of human O-GlcNAcase show distinct catalytic efficiencies". Biochemistry. Biokhimiia. 75 (7): 938–43. doi:10.1134/S0006297910070175. PMID 20673219.
- Greig, Ian; Vocadlo, David. "Glycoside Hydrolase Family 84". Cazypedia. Retrieved 28 March 2017.
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW (March 2001). "Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain". The Journal of Biological Chemistry. 276 (13): 9838–45. doi:10.1074/jbc.M010420200. PMID 11148210.
- Lima VV, Rigsby CS, Hardy DM, Webb RC, Tostes RC (2009). "O-GlcNAcylation: a novel post-translational mechanism to alter vascular cellular signaling in health and disease: focus on hypertension". Journal of the American Society of Hypertension. 3 (6): 374–87. doi:10.1016/j.jash.2009.09.004. PMC 3022480. PMID 20409980.
- Förster S, Welleford AS, Triplett JC, Sultana R, Schmitz B, Butterfield DA (September 2014). "Increased O-GlcNAc levels correlate with decreased O-GlcNAcase levels in Alzheimer disease brain". Biochimica et Biophysica Acta. 1842 (9): 1333–9. doi:10.1016/j.bbadis.2014.05.014. PMC 4140188. PMID 24859566.
- Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, et al. (May 2000). "The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny". Proceedings of the National Academy of Sciences of the United States of America. 97 (11): 5735–9. doi:10.1073/pnas.100471497. PMC 18502. PMID 10801981.
- Love DC, Ghosh S, Mondoux MA, Fukushige T, Wang P, Wilson MA, et al. (April 2010). "Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity". Proceedings of the National Academy of Sciences of the United States of America. 107 (16): 7413–8. doi:10.1073/pnas.0911857107. PMC 2867743. PMID 20368426.
- Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, et al. (April 2006). "Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity". Nature Structural & Molecular Biology. 13 (4): 365–71. doi:10.1038/nsmb1079. PMID 16565725.
- Alonso J, Schimpl M, van Aalten DM (December 2014). "O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling?". The Journal of Biological Chemistry. 289 (50): 34433–9. doi:10.1074/jbc.R114.609198. PMC 4263850. PMID 25336650.
- Lim S, Haque MM, Nam G, Ryoo N, Rhim H, Kim YK (August 2015). "Monitoring of Intracellular Tau Aggregation Regulated by OGA/OGT Inhibitors". International Journal of Molecular Sciences. 16 (9): 20212–24. doi:10.3390/ijms160920212. PMC 4613198. PMID 26343633.
- Roth C, Chan S, Offen WA, Hemsworth GR, Willems LI, King DT, et al. (June 2017). "Structural and functional insight into human O-GlcNAcase". Nature Chemical Biology. 13 (6): 610–612. doi:10.1038/nchembio.2358. PMC 5438047. PMID 28346405.
Further reading
- Nakajima D, Okazaki N, Yamakawa H, Kikuno R, Ohara O, Nagase T (June 2002). "Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones". DNA Research. 9 (3): 99–106. doi:10.1093/dnares/9.3.99. PMID 12168954.
- Ishikawa K, Nagase T, Suyama M, Miyajima N, Tanaka A, Kotani H, et al. (June 1998). "Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro". DNA Research. 5 (3): 169–76. doi:10.1093/dnares/5.3.169. PMID 9734811.
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW (March 2001). "Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain". The Journal of Biological Chemistry. 276 (13): 9838–45. doi:10.1074/jbc.M010420200. PMID 11148210.
- Comtesse N, Maldener E, Meese E (May 2001). "Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a beta-N-acetylglucosaminidase". Biochemical and Biophysical Research Communications. 283 (3): 634–40. doi:10.1006/bbrc.2001.4815. PMID 11341771.
- Wells L, Gao Y, Mahoney JA, Vosseller K, Chen C, Rosen A, Hart GW (January 2002). "Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase". The Journal of Biological Chemistry. 277 (3): 1755–61. doi:10.1074/jbc.M109656200. PMID 11788610.
- Farook VS, Bogardus C, Prochazka M (2003). "Analysis of MGEA5 on 10q24.1-q24.3 encoding the beta-O-linked N-acetylglucosaminidase as a candidate gene for type 2 diabetes mellitus in Pima Indians" (PDF). Molecular Genetics and Metabolism. 77 (1–2): 189–93. doi:10.1016/S1096-7192(02)00127-0. PMID 12359146.
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villén J, Li J, et al. (August 2004). "Large-scale characterization of HeLa cell nuclear phosphoproteins". Proceedings of the National Academy of Sciences of the United States of America. 101 (33): 12130–5. doi:10.1073/pnas.0404720101. PMC 514446. PMID 15302935.
- Ballif BA, Villén J, Beausoleil SA, Schwartz D, Gygi SP (November 2004). "Phosphoproteomic analysis of the developing mouse brain". Molecular & Cellular Proteomics. 3 (11): 1093–101. doi:10.1074/mcp.M400085-MCP200. PMID 15345747.
- Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE (December 2004). "Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities". The Journal of Biological Chemistry. 279 (51): 53665–73. doi:10.1074/jbc.M410406200. PMID 15485860.
- Whisenhunt TR, Yang X, Bowe DB, Paterson AJ, Van Tine BA, Kudlow JE (June 2006). "Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development". Glycobiology. 16 (6): 551–63. doi:10.1093/glycob/cwj096. PMID 16505006.
- Toleman C, Paterson AJ, Kudlow JE (May 2006). "Location and characterization of the O-GlcNAcase active site". Biochimica et Biophysica Acta. 1760 (5): 829–39. doi:10.1016/j.bbagen.2006.01.017. PMID 16517082.
- Cameron EA, Martinez-Marignac VL, Chan A, Valladares A, Simmonds LV, Wacher N, et al. (2007). "MGEA5-14 polymorphism and type 2 diabetes in Mexico City". American Journal of Human Biology. 19 (4): 593–6. doi:10.1002/ajhb.20639. PMID 17546623.
External links
- Protein+O-GlcNAcase at the US National Library of Medicine Medical Subject Headings (MeSH)