Photosystem I
| Photosystem I | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Plant photosystem I, PDB 2001 | |||||||||
| Identifiers | |||||||||
| EC number | 1.97.1.12 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
| PsaA_PsaB | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of photosystem I: a photosynthetic reaction center and core antenna system from cyanobacteria | |||||||||
| Identifiers | |||||||||
| Symbol | PsaA_PsaB | ||||||||
| Pfam | PF00223 | ||||||||
| InterPro | IPR001280 | ||||||||
| PROSITE | PDOC00347 | ||||||||
| SCOP | 1jb0 | ||||||||
| SUPERFAMILY | 1jb0 | ||||||||
| TCDB | 5.B.4 | ||||||||
| OPM superfamily | 2 | ||||||||
| OPM protein | 1jb0 | ||||||||
| Membranome | 535 | ||||||||
| |||||||||
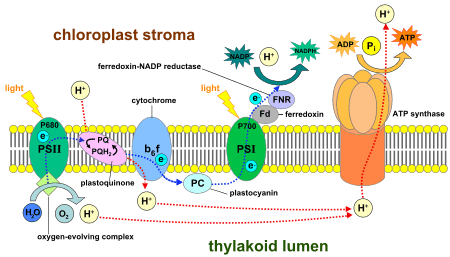
Photosystem I (PS I, or plastocyanin-ferredoxin oxidoreductase) is the second photosystem in the photosynthetic light reactions of algae, plants, and some bacteria. Photosystem I (PS I) [1] is an integral membrane protein complex that uses light energy to produce the high energy carriers ATP and NADPH.[2] More than 110 cofactors, significantly more than photosystem II, comprise the PS I system.[3]
History
Photosystem I is named because it was discovered before photosystem II. Aspects of PS I were discovered in the 1950s, but the significances of these discoveries was not yet known.[4] Louis Duysens first proposed the concepts of photosystems I and II in 1960, and, in the same year, a proposal by Fay Bendall and Robert Hill assembled earlier discoveries into a cohesive theory of serial photosynthetic reactions.[4] Hill and Bendall's hypothesis was later justified in experiments conducted in 1961 by Duysens and Witt groups.[4]
Components and action of photosystem I
Two main subunits of PS I, PsaA and PsaB, are closely related proteins involved in the binding of P700, A0, A1, and Fx. PsaA and PsaB are both integral membrane proteins of 730 to 750 amino acids that seem to contain 11 transmembrane segments. The Fx 4Fe-4S iron-sulphur centre is bound by four cysteines; two of these cysteines are provided by the PsaA protein and the two others by PsaB. The two cysteines in both proteins are proximal and located in a loop between the ninth and tenth transmembrane segments. A leucine zipper motif seems to be present [5] downstream of the cysteines and could contribute to dimerisation of psaA/psaB. The terminal electron acceptors, FA and FB, are located in a 9 kDa protein called PsaC.[6][7]
| Protein subunits | Proton turn into form of ATP |
|---|---|
| Subunit | Description |
| PsaA | |
| PsaB | |
| PsaC | |
| PsaD | |
| PsaE | |
| PsaI | |
| PsaJ | |
| PsaK | |
| PsaL | |
| PsaM | |
| PsaX | |
| Cytochrome b6f complex | Soluble protein |
| Fa | In electron transport chain (ETC) |
| Fb | In ETC |
| Fx | In ETC |
| Ferredoxin | Electron carrier in ETC |
| Plastocyanin | Soluble protein |
| Lipids | |
| MGDG II | Monogalactosyldiglyceride lipid |
| PG I | Phosphatidylglycerol phospholipid |
| PG III | Phosphatidylglycerol phospholipid |
| PG IV | Phosphatidylglycerol phospholipid |
| Pigments | |
| Chlorophyll a | 90 pigment molecules in antenna system |
| Chlorophyll a | 5 pigment molecules in ETC |
| Chlorophyll a0 | Early electron acceptor of modified chlorophyll in ETC |
| Chlorophyll a′ | 1 pigment molecule in ETC |
| β-Carotene | 22 carotenoid pigment molecules |
| Coenzymes and cofactors | |
| Molecule | Description |
| QK-A | Early electron acceptor vitamin K1 phylloquinone in ETC |
| QK-B | Early electron acceptor vitamin K1 phylloquinone in ETC |
| FNR | Ferredoxin-NADP+ oxidoreductase enzyme |
| Ca2+ |
Calcium ion |
| Mg2+ |
Magnesium ion |
Photon
Photoexcitation of the pigment molecules in the antenna complex induces electron transfer.[9]
Antenna complex
The antenna complex is composed of molecules of chlorophyll and carotenoids mounted on two proteins.[10] These pigment molecules transmit the resonance energy from photons when they become photoexcited. Antenna molecules can absorb all wavelengths of light within the visible spectrum.[11] The number of these pigment molecules varies from organism to organism. For instance, the cyanobacterium Synechococcus elongatus (Thermosynechococcus elongatus) has about 100 chlorophylls and 20 carotenoids, whereas spinach chloroplasts have around 200 chlorophylls and 50 carotenoids.[11][3] Located within the antenna complex of PS I are molecules of chlorophyll called P700 reaction centers. The energy passed around by antenna molecules is directed to the reaction center. There may be as many as 120 or as few as 25 chlorophyll molecules per P700.[12]
P700 reaction center
The P700 reaction center is composed of modified chlorophyll a that best absorbs light at a wavelength of 700 nm, with higher wavelengths causing bleaching.[13] P700 receives energy from antenna molecules and uses the energy from each photon to raise an electron to a higher energy level. These electrons are moved in pairs in an oxidation/reduction process from P700 to electron acceptors. P700 has an electric potential of about −1.2 volts. The reaction center is made of two chlorophyll molecules and is therefore referred to as a dimer.[10] The dimer is thought to be composed of one chlorophyll a molecule and one chlorophyll a′ molecule (P700, webber). However, if P700 forms a complex with other antenna molecules, it can no longer be a dimer.[12]
Modified chlorophyll a0
Modified chlorophyll a0 is an early electron acceptor in PS I. Chlorophyll a0 accepts electrons from P700 before passing them along to another early electron acceptor.[13]
Phylloquinone A1
Phylloquinone A1 is the next early electron acceptor in PS I. Phylloquinone is also sometimes called vitamin K1.[14] Phylloquinone A1 oxidizes A0 in order to receive the electron and in turn reduces Fx in order to pass the electron to Fb and Fa.[14][15]
Iron–sulfur complex
Three proteinaceous iron–sulfur reaction centers are found in PS I. Labeled Fx, Fa, and Fb, they serve as electron relays.[16] Fa and Fb are bound to protein subunits of the PS I complex and Fx is tied to the PS I complex.[16] Various experiments have shown some disparity between theories of iron–sulfur cofactor orientation and operation order.[16]
Ferredoxin
Ferredoxin (Fd) is a soluble protein that facilitates reduction of NADP+
to NADPH.[17] Fd moves to carry an electron either to a lone thylakoid or to an enzyme that reduces NADP+
.[17] Thylakoid membranes have one binding site for each function of Fd.[17] The main function of Fd is to carry an electron from the iron-sulfur complex to the enzyme ferredoxin–NADP+
reductase.[17]
Ferredoxin–NADP+
reductase (FNR)
This enzyme transfers the electron from reduced ferredoxin to NADP+
to complete the reduction to NADPH.[18] FNR may also accept an electron from NADPH by binding to it.[18]
Plastocyanin
Plastocyanin is an electron carrier that transfers the electron from cytochrome b6f to the P700 cofactor of PS I.[9][19]
Ycf4 protein domain
The Ycf4 protein domain is found on the thylakoid membrane and is vital to photosystem I. This thylakoid transmembrane protein helps assemble the components of photosystem I, without it, photosynthesis would be inefficient.[20]
Green sulfur bacteria and the evolution of PS I
Molecular data show that PS I likely evolved from the photosystems of green sulfur bacteria. The photosystems of green sulfur bacteria and those of cyanobacteria, algae, and higher plants are not the same, however there are many analogous functions and similar structures. Three main features are similar between the different photosystems.[21] First, redox potential is negative enough to reduce ferredoxin.[21] Next, the electron-accepting reaction centers include iron–sulfur proteins.[21] Last, redox centres in complexes of both photosystems are constructed upon a protein subunit dimer.[21] The photosystem of green sulfur bacteria even contains all of the same cofactors of the electron transport chain in PS I.[21] The number and degree of similarities between the two photosystems strongly indicates that PS I is derived from the analogous photosystem of green sulfur bacteria.
See also
References
- ↑ Golbeck JH (1987). "Structure, function and organization of the Photosystem I reaction center complex". Biochimica et Biophysica Acta. 895 (3): 167–204. doi:10.1016/s0304-4173(87)80002-2. PMID 3333014.
- ↑ Yamori W, Shikanai T (April 2016). "Physiological Functions of Cyclic Electron Transport Around Photosystem I in Sustaining Photosynthesis and Plant Growth". Annual Review of Plant Biology. 67: 81–106. doi:10.1146/annurev-arplant-043015-112002. PMID 26927905.
- 1 2 Nelson N, Yocum CF (2006). "Structure and function of photosystems I and II". Annual Review of Plant Biology. 57: 521–65. doi:10.1146/annurev.arplant.57.032905.105350. PMID 16669773.
- 1 2 3 Fromme P, Mathis P (2004). "Unraveling the photosystem I reaction center: a history, or the sum of many efforts". Photosynthesis Research. 80 (1–3): 109–24. doi:10.1023/B:PRES.0000030657.88242.e1. PMID 16328814.
- ↑ Webber AN, Malkin R (May 1990). "Photosystem I reaction-centre proteins contain leucine zipper motifs. A proposed role in dimer formation". FEBS Letters. 264 (1): 1–4. doi:10.1016/0014-5793(90)80749-9. PMID 2186925.
- ↑ Jagannathan B, Golbeck JH (April 2009). "Breaking biological symmetry in membrane proteins: the asymmetrical orientation of PsaC on the pseudo-C2 symmetric Photosystem I core". Cellular and Molecular Life Sciences. 66 (7): 1257–70. doi:10.1007/s00018-009-8673-x. PMID 19132290.
- ↑ Jagannathan B, Golbeck JH (June 2009). "Understanding of the binding interface between PsaC and the PsaA/PsaB heterodimer in photosystem I". Biochemistry. 48 (23): 5405–16. doi:10.1021/bi900243f. PMID 19432395.
- ↑ Saenger W, Jordan P, Krauss N (April 2002). "The assembly of protein subunits and cofactors in photosystem I". Current Opinion in Structural Biology. 12 (2): 244–54. doi:10.1016/S0959-440X(02)00317-2. PMID 11959504.
- 1 2 Raven PH, Evert RF, Eichhorn SE (2005). "Photosynthesis, Light, and Life". Biology of Plants (7th ed.). New York: W. H. Freeman. pp. 121–127. ISBN 978-0-7167-1007-3.
- 1 2 Zeiger E, Taiz L (2006). "Ch. 7: Topic 7.8: Photosystem I". Plant Physiology (4th ed.). Sunderland, MA: Sinauer Associates. ISBN 0-87893-856-7.
- 1 2 "The Photosynthetic Process". Archived from the original on 2009-02-19.
- 1 2 Shubin VV, Karapetyan NV, Krasnovsky AA (January 1986). "Molecular arrangement of pigment-protein complex of photosystem 1". Photosynthesis Research. 9 (1–2): 3–12. doi:10.1007/BF00029726. PMID 24442279.
- 1 2 Rutherford AW, Heathcote P (December 1985). "Primary photochemistry in photosystem-I". Photosynthesis Research. 6 (4): 295–316. doi:10.1007/BF00054105. PMID 24442951.
- 1 2 Itoh S, Iwaki M (1989). "Vitamin K1 (Phylloquinone) Restores the Turnover of FeS centers of Ether-extracted Spinach PS I Particles". FEBS Letters. 243 (1): 47–52. doi:10.1016/0014-5793(89)81215-3.
- ↑ Palace GP, Franke JE, Warden JT (May 1987). "Is phylloquinone an obligate electron carrier in photosystem I?". FEBS Letters. 215 (1): 58–62. doi:10.1016/0014-5793(87)80113-8. PMID 3552735.
- 1 2 3 Vassiliev IR, Antonkine ML, Golbeck JH (October 2001). "Iron-sulfur clusters in type I reaction centers". Biochimica et Biophysica Acta. 1507 (1–3): 139–60. doi:10.1016/S0005-2728(01)00197-9. PMID 11687212.
- 1 2 3 4 Forti G, Maria P, Grubas G (1985). "Two Sites of Interaction of Ferredoxin with thylakoids". FEBS Letters. 186 (2): 149–152. doi:10.1016/0014-5793(85)80698-0.
- 1 2 Madoz J, Fernández Recio J, Gómez Moreno C, Fernández VM (November 1998). "Investigation of the Diaphorase Reaction of Ferredoxin–NADP+
Reductase by Electrochemical Methods" (PDF). Bioelectrochemistry and Bioenergetics. 47 (1): 179–183. doi:10.1016/S0302-4598(98)00175-5. - ↑ Hope AB (January 2000). "Electron transfers amongst cytochrome f, plastocyanin and photosystem I: kinetics and mechanisms". Biochimica et Biophysica Acta. 1456 (1): 5–26. doi:10.1016/S0005-2728(99)00101-2. PMID 10611452.
- ↑ Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD (October 1997). "The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex". The EMBO Journal. 16 (20): 6095–104. doi:10.1093/emboj/16.20.6095. PMC 1326293. PMID 9321389.
- 1 2 3 4 5 Lockau W, Nitschke W (1993). "Photosystem I and its Bacterial Counterparts". Physiologia Plantarum. 88 (2): 372–381. doi:10.1111/j.1399-3054.1993.tb05512.x.