HEXB
Beta-hexosaminidase subunit beta is an enzyme that in humans is encoded by the HEXB gene.[5][6][7]
Hexosaminidase B is the beta subunit of the lysosomal enzyme beta-hexosaminidase that, together with the cofactor GM2 activator protein, catalyzes the degradation of the ganglioside GM2, and other molecules containing terminal N-acetyl hexosamines. Beta-hexosaminidase is composed of two subunits, alpha and beta, which are encoded by separate genes. Both beta-hexosaminidase alpha and beta subunits are members of family 20 of glycosyl hydrolases. Mutations in the alpha or beta subunit genes lead to an accumulation of GM2 ganglioside in neurons and neurodegenerative disorders termed the GM2 gangliosidoses. Beta subunit gene mutations lead to Sandhoff disease (GM2-gangliosidosis type II).[7]
Structure
Gene
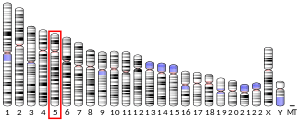
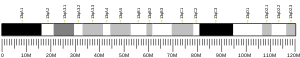
The HEXB gene lies on the chromosome location of 5q13.3 and consists of 15 exons, spanning 35-40Kb.
Protein
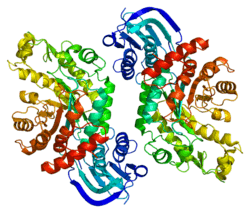
HEXB consists of 556 amino acid residues and weighs 63111Da.
Function
HEXB is one of the two subunits forming β-hexosaminidase which functions as a glycosyl hydrolase that remove β-linked nonreducing-terminal GalNAc or GlcNAc residues in the lysosome.[8] Inability of HEXB will lead toβ-hexosaminidase defect and result in a group of recessive disorders called GM2 gangliosidoses, characterized by the accumulation of GM2 ganglioside.[9]
Clinical significance
Genetic defects in HEXB can result in the accumulation of GM2 ganglioside in neural tissues and two of three lysosomal storage diseases collectively known as GM2 gangliosidosis, of which Sandhoff disease (defects in the β subunit) is the best studied one.[8] Patients present with neurosomatic manifestations. Therapeutic effects of Hex subunit gene transduction have been examined on Sandhoff disease model mice.[10] Intracerebroventricular administration of the modified β-hexosaminidase B to Sandhoff mode mice restored the β-hexosaminidase activity in the brains, and reduced the GM2 ganglioside storage in the parenchyma.[11]
Interactions
HEXB has been found to interact with HEXA[12] and ganglioside.[10]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000049860 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000021665 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ O'Dowd BF, Quan F, Willard HF, Lamhonwah AM, Korneluk RG, Lowden JA, Gravel RA, Mahuran DJ (February 1985). "Isolation of cDNA clones coding for the beta subunit of human beta-hexosaminidase". Proceedings of the National Academy of Sciences of the United States of America. 82 (4): 1184–8. doi:10.1073/pnas.82.4.1184. PMC 397219. PMID 2579389.
- ↑ Korneluk RG, Mahuran DJ, Neote K, Klavins MH, O'Dowd BF, Tropak M, Willard HF, Anderson MJ, Lowden JA, Gravel RA (June 1986). "Isolation of cDNA clones coding for the alpha-subunit of human beta-hexosaminidase. Extensive homology between the alpha- and beta-subunits and studies on Tay-Sachs disease". The Journal of Biological Chemistry. 261 (18): 8407–13. PMID 3013851.
- 1 2 "Entrez Gene: HEXB hexosaminidase B (beta polypeptide)".
- 1 2 Bateman KS, Cherney MM, Mahuran DJ, Tropak M, James MN (March 2011). "Crystal structure of β-hexosaminidase B in complex with pyrimethamine, a potential pharmacological chaperone". Journal of Medicinal Chemistry. 54 (5): 1421–9. doi:10.1021/jm101443u. PMC 3201983. PMID 21265544.
- ↑ Sonnino S, Chigorno V (September 2000). "Ganglioside molecular species containing C18- and C20-sphingosine in mammalian nervous tissues and neuronal cell cultures". Biochimica et Biophysica Acta. 1469 (2): 63–77. doi:10.1016/s0005-2736(00)00210-8. PMID 10998569.
- 1 2 Itakura T, Kuroki A, Ishibashi Y, Tsuji D, Kawashita E, Higashine Y, Sakuraba H, Yamanaka S, Itoh K (August 2006). "Inefficiency in GM2 ganglioside elimination by human lysosomal beta-hexosaminidase beta-subunit gene transfer to fibroblastic cell line derived from Sandhoff disease model mice". Biological & Pharmaceutical Bulletin. 29 (8): 1564–9. doi:10.1248/bpb.29.1564. PMID 16880605.
- ↑ Matsuoka K, Tamura T, Tsuji D, Dohzono Y, Kitakaze K, Ohno K, Saito S, Sakuraba H, Itoh K (June 2011). "Therapeutic potential of intracerebroventricular replacement of modified human β-hexosaminidase B for GM2 gangliosidosis". Molecular Therapy. 19 (6): 1017–24. doi:10.1038/mt.2011.27. PMC 3129794. PMID 21487393.
- ↑ Gort L, de Olano N, Macías-Vidal J, Coll MA (September 2012). "GM2 gangliosidoses in Spain: analysis of the HEXA and HEXB genes in 34 Tay-Sachs and 14 Sandhoff patients". Gene. 506 (1): 25–30. doi:10.1016/j.gene.2012.06.080. PMID 22789865.
Further reading
- Mahuran DJ (February 1991). "The biochemistry of HEXA and HEXB gene mutations causing GM2 gangliosidosis". Biochimica et Biophysica Acta. 1096 (2): 87–94. doi:10.1016/0925-4439(91)90044-A. PMID 1825792.
- Mahuran DJ (October 1999). "Biochemical consequences of mutations causing the GM2 gangliosidoses". Biochimica et Biophysica Acta. 1455 (2–3): 105–38. doi:10.1016/S0925-4439(99)00074-5. PMID 10571007.
- Gilbert F, Kucherlapati R, Creagan RP, Murnane MJ, Darlington GJ, Ruddle FH (January 1975). "Tay-Sachs' and Sandhoff's diseases: the assignment of genes for hexosaminidase A and B to individual human chromosomes". Proceedings of the National Academy of Sciences of the United States of America. 72 (1): 263–7. doi:10.1073/pnas.72.1.263. PMC 432284. PMID 1054503.
- McInnes B, Potier M, Wakamatsu N, Melancon SB, Klavins MH, Tsuji S, Mahuran DJ (August 1992). "An unusual splicing mutation in the HEXB gene is associated with dramatically different phenotypes in patients from different racial backgrounds". The Journal of Clinical Investigation. 90 (2): 306–14. doi:10.1172/JCI115863. PMC 443103. PMID 1386607.
- Bolhuis PA, Bikker H (November 1992). "Deletion of the 5'-region in one or two alleles of HEXB in 15 out of 30 patients with Sandhoff disease". Human Genetics. 90 (3): 328–9. doi:10.1007/bf00220096. PMID 1487253.
- Wakamatsu N, Kobayashi H, Miyatake T, Tsuji S (February 1992). "A novel exon mutation in the human beta-hexosaminidase beta subunit gene affects 3' splice site selection". The Journal of Biological Chemistry. 267 (4): 2406–13. PMID 1531140.
- Banerjee P, Siciliano L, Oliveri D, McCabe NR, Boyers MJ, Horwitz AL, Li SC, Dawson G (November 1991). "Molecular basis of an adult form of beta-hexosaminidase B deficiency with motor neuron disease". Biochemical and Biophysical Research Communications. 181 (1): 108–15. doi:10.1016/S0006-291X(05)81388-9. PMID 1720305.
- Boose JA, Tifft CJ, Proia RL, Myerowitz R (November 1990). "Synthesis of a human lysosomal enzyme, beta-hexosaminidase B, using the baculovirus expression system". Protein Expression and Purification. 1 (2): 111–20. doi:10.1016/1046-5928(90)90003-H. PMID 1967020.
- Mahuran DJ (April 1990). "Characterization of human placental beta-hexosaminidase I2. Proteolytic processing intermediates of hexosaminidase A". The Journal of Biological Chemistry. 265 (12): 6794–9. PMID 2139028.
- Neote K, McInnes B, Mahuran DJ, Gravel RA (November 1990). "Structure and distribution of an Alu-type deletion mutation in Sandhoff disease". The Journal of Clinical Investigation. 86 (5): 1524–31. doi:10.1172/JCI114871. PMC 296899. PMID 2147027.
- Neote K, Brown CA, Mahuran DJ, Gravel RA (December 1990). "Translation initiation in the HEXB gene encoding the beta-subunit of human beta-hexosaminidase". The Journal of Biological Chemistry. 265 (34): 20799–806. PMID 2147427.
- Dlott B, d'Azzo A, Quon DV, Neufeld EF (October 1990). "Two mutations produce intron insertion in mRNA and elongated beta-subunit of human beta-hexosaminidase". The Journal of Biological Chemistry. 265 (29): 17921–7. PMID 2170400.
- Nakano T, Suzuki K (March 1989). "Genetic cause of a juvenile form of Sandhoff disease. Abnormal splicing of beta-hexosaminidase beta chain gene transcript due to a point mutation within intron 12". The Journal of Biological Chemistry. 264 (9): 5155–8. PMID 2522450.
- Hubbes M, Callahan J, Gravel R, Mahuran D (June 1989). "The amino-terminal sequences in the pro-alpha and -beta polypeptides of human lysosomal beta-hexosaminidase A and B are retained in the mature isozymes". FEBS Letters. 249 (2): 316–20. doi:10.1016/0014-5793(89)80649-0. PMID 2525487.
- Bikker H, van den Berg FM, Wolterman RA, de Vijlder JJ, Bolhuis PA (February 1989). "Demonstration of a Sandhoff disease-associated autosomal 50-kb deletion by field inversion gel electrophoresis". Human Genetics. 81 (3): 287–8. doi:10.1007/BF00279006. PMID 2921040.
- Bolhuis PA, Oonk JG, Kamp PE, Ris AJ, Michalski JC, Overdijk B, Reuser AJ (January 1987). "Ganglioside storage, hexosaminidase lability, and urinary oligosaccharides in adult Sandhoff's disease". Neurology. 37 (1): 75–81. doi:10.1212/wnl.37.1.75. PMID 2948136.
- Proia RL (March 1988). "Gene encoding the human beta-hexosaminidase beta chain: extensive homology of intron placement in the alpha- and beta-chain genes". Proceedings of the National Academy of Sciences of the United States of America. 85 (6): 1883–7. doi:10.1073/pnas.85.6.1883. PMC 279885. PMID 2964638.
- Mahuran DJ, Neote K, Klavins MH, Leung A, Gravel RA (April 1988). "Proteolytic processing of pro-alpha and pro-beta precursors from human beta-hexosaminidase. Generation of the mature alpha and beta a beta b subunits". The Journal of Biological Chemistry. 263 (10): 4612–8. PMID 2965147.