Deletion (genetics)
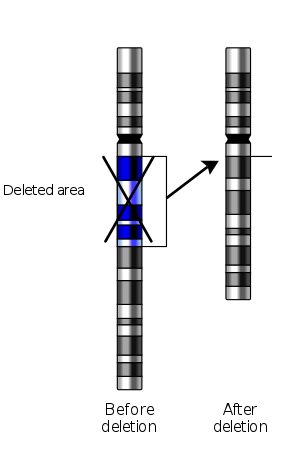
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is lost during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome.[1] The smallest single base deletion mutations are believed to occur by a single base flipping in the template DNA, followed by template DNA strand slippage, within the DNA polymerase active site.[2][3][4] Deletions can be caused by errors in chromosomal crossover during meiosis, which causes several serious genetic diseases. Deletions that do not occur in multiples of three bases can cause a frameshift by changing the 3-nucleotide protein reading frame of the genetic sequence. The examples are given below of types and effects of deletions are representative of eukaryotic organisms, particularly humans, but are not relevant to prokaryotic organisms such as bacteria.
Causes
Causes include the following:
- Losses from translocation
- Chromosomal crossovers within a chromosomal inversion
- Unequal crossing over
- Breaking without rejoining
For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop.
Types
Types of deletion include the following:
- 'Terminal Deletion' — a deletion that occurs towards the end of a chromosome.
- Intercalary Deletion / Interstitial Addition — a deletion that occurs from the interior of a chromosome.
- Microdeletion — a relatively small amount of deletion (up to 5Mb that could include a dozen genes).
Microdeletion is usually found in children with physical abnormalities. A large amount of deletion would result in immediate abortion (miscarriage).
Effects
Small deletions are less likely to be fatal; large deletions are usually fatal — there are always variations based on which genes are lost. Some medium-sized deletions lead to recognizable human disorders, e.g. Williams syndrome.
Deletion of a number of pairs that is not evenly divisible by three will lead to a frameshift mutation, causing all of the codons occurring after the deletion to be read incorrectly during translation, producing a severely altered and potentially nonfunctional protein. In contrast, a deletion that is evenly divisible by three is called an in-frame deletion.[5]
Deletions are responsible for an array of genetic disorders, including some cases of male infertility and two thirds of cases of Duchenne muscular dystrophy.[1] Deletion of part of the short arm of chromosome 5 results in Cri du chat syndrome.[1] Deletions in the SMN-encoding gene cause spinal muscular atrophy, the most common genetic cause of infant death.
Recent work suggests that some deletions of highly conserved sequences (CONDELs) may be responsible for the evolutionary differences present among closely related species. Such deletions in humans are referred to as hCONDELs may be responsible for the anatomical and behavioral differences between humans, chimpanzees and other mammals.[6]
Detection
The introduction of molecular techniques in conjunction with classical cytogenetic methods has in recent years greatly improved the diagnostic potential for chromosomal abnormalities. In particular, microarray-comparative genomic hybridization (CGH) based on the use of BAC clones promises a sensitive strategy for the detection of DNA copy-number changes on a genome-wide scale. The resolution of detection could be as high as >30,000 "bands" and the size of chromosomal deletion detected could as small as 5–20 kb in length.[7] Other computation methods were selected to discover DNA sequencing deletion errors such as end-sequence profiling.[8][9]
See also
References
- 1 2 3 Lewis, R. (2004). Human Genetics: Concepts and Applications (6th ed.). McGraw Hill. ISBN 0072951745.
- ↑ Banavali, Nilesh K. "Partial base flipping is sufficient for strand slippage near DNA duplex termini." Journal of the American Chemical Society, 2013, 135(22), 8274-8282.
- ↑ Banavali, Nilesh K. "Analyzing the Relationship between Single Base Flipping and Strand Slippage near DNA Duplex Termini." Journal of Physical Chemistry B, 2013, 117(46), 14320-14328.
- ↑ Manjari, Swati R., Pata, Janice D., Banavali, Nilesh K. "Cytosine Unstacking and Strand Slippage at an Insertion–Deletion Mutation Sequence in an Overhang-Containing DNA Duplex." Biochemistry, 2014, 53(23), 3807-3816.
- ↑ LSDB — Controlled vocabulary terms at The GEN2PHEN Knowledge Centre. Posted Fri, 08/01/2010.
- ↑ McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, Wenger AM, Bejerano G, Kingsley DM (March 2011). "Human-specific loss of regulatory DNA and the evolution of human-specific traits". Nature. 471 (7337): 216–9. doi:10.1038/nature09774. PMC 3071156. PMID 21390129.
- ↑ Ren, H (May 2005). "BAC-based PCR fragment microarray: high-resolution detection of chromosomal deletion and duplication breakpoints". Human Mutation. 25 (5): 476–482. doi:10.1002/humu.20164. PMID 15832308.
- ↑ Shmilovici, A.; Ben-Gal, I. (2007). "Using a VOM Model for Reconstructing Potential Coding Regions in EST Sequences" (PDF). Journal of Computational Statistics. 22 (1): 49–69. doi:10.1007/s00180-007-0021-8.
- ↑ Volik, S.; Zhao, S.; Chin, K.; Brebner, J. H.; Herndon, D. R.; Tao, Q.; Kowbel, D.; Huang, G.; Lapuk, A.; Kuo, W.-L.; Magrane, G.; de Jong, P.; Gray, J. W.; Collins, C. (4 June 2003). "End-sequence profiling: Sequence-based analysis of aberrant genomes". Proceedings of the National Academy of Sciences. 100 (13): 7696–7701. doi:10.1073/pnas.1232418100. PMC 164650. PMID 12788976.
| Wikimedia Commons has media related to Deletion (genetics). |