Citrus taxonomy

The botanical classification of the species, hybrids, varieties and cultivars belonging to the genus Citrus is called "citrus taxonomy".
Citrus taxonomy refers to the botanical classification of the species, varieties, cultivars, and graft hybrids within the genus Citrus and related genera, found in cultivation and in the wild.
Citrus taxonomy is complex.[1] Cultivated citrus are derived from various citrus species found in the wild. Some are only selections of the original wild types, while others are hybrids between two or more ancestors. Citrus plants hybridize easily between species with completely different morphologies, and similar-looking citrus fruits may have quite different ancestries.[2][3] Some differ only in disease resistance.[4] Conversely, different-looking varieties may be nearly genetically identical, and differ only by a bud mutation.[5]
Detailed genomic analysis of wild and domesticated citrus cultivars has suggested that the progenitor of modern citrus species expanded out of the Himalayan foothills in a rapid radiation that has produced at least 10 wild species in South and East Asia and Australia. Most commercial cultivars are the product of hybridization among these wild species, with most coming from crosses involving citrons, mandarins and pomelos.[6] Many different phylogenies for the non-hybrid citrus have been proposed,[7] and the phylogeny based on their nuclear genome does not match that derived from their chloroplast DNA, probably a consequence of the rapid initial divergence.[6] Taxonomic terminology is not yet settled.
Most hybrids express different ancestral traits when planted from seeds (F2 hybrids) and can continue a stable lineage only through vegetative propagation. Some hybrids do reproduce true to type via nucellar seeds in a process called apomixis.[3] As such, many hybrid species represent the clonal progeny of a single original F1 cross, though others combine fruit with similar characteristics that have arisen from distinct crosses.
Genetic history

All of the wild 'pure' citrus species trace to a common ancestor that lived in the Himalayan foothills, where a late-Miocene citrus fossil, Citrus linczangensis, has been found. At that time, a lessening of the monsoons and resultant drier climate in the region allowed the citrus ancestor to expand across south and east Asia in a rapid genetic radiation. After the plant crossed the Wallace line a second radiation took place in the early Pliocene (about 4 million years ago) to give rise to the Australian species. Most modern cultivars are actually hybrids derived from a small number of 'pure' original species. Though hundreds of species names have been assigned, a recent genomic study by Wu, et al. identified just ten ancestral species of citrus among those studied. Of these, the seven were native to Asia: pomelo (Citrus maxima), the 'pure' mandarins (C. reticulata - most mandarin cultivars were hybrids of this species with pomelo), citrons (C. medica), micranthas (C. micrantha), the Ichang papeda (C. ichangensis), the mangshanyegan (C. mangshanensis), and the oval (Nagami) kumquat (Fortunella margarita or C. japonica var. margarita). In Australia there were three identified, the desert lime (C. glauca), round lime (C. australis) and the finger lime (C. australasica). Many other cultivars previously identified as species were found to be closely related variants (subspecies or varieties) or hybrids of these species,[6] though not all cultivars were evaluated.
Interbreeding seems possible between all citrus plants, and between citrus plants and some plants which may or may not be categorized as citrus. The ability of citrus hybrids to self-pollinate and to reproduce sexually also helps create new varieties. The three predominant ancestral citrus taxa are citron (C. medica), pomelo (C. maxima), and mandarin (C. reticulata).[6] These taxa interbreed freely, despite being quite genetically distinct, having arisen through allopatric speciation, with citrons evolving in northern Indochina, pomelos in the Malay Archipelago, and mandarins in Vietnam, southern China, and Japan.[9] The hybrids of these taxa include familiar citrus fruits like oranges, grapefruit, lemons, limes, and some tangerines.[1][10] Citrons have also been hybridized with other citrus taxa, for example, crossed with micrantha to produce the Key lime.[6] In many cases, these crops are propagated asexually, and lose their characteristic traits if bred. However, some of these hybrids have interbred with one another and with the original taxa, making the citrus family tree a complicated network.
Kumquats do not naturally interbreed with core taxa due to different flowering times,[11] but hybrids (such as the calamondin) exist. Australian limes are native to Australia and Papua New Guinea, so they did not naturally interbreed with the core taxa, but they have been crossbred with mandarins and calamondins by modern breeders. Humans have deliberately bred new citrus fruits by propagating seedlings of spontaneous crosses (e.g. clementines), creating or selecting mutations of hybrids, (e.g. Meyer lemon), and crossing different varieties (e.g. 'Australian Sunrise', a finger lime and calamondin cross).
Citrus naming systems
Initially, many citrus types were identified and named by individual taxonomists, resulting in a large number of identified species: 870 by a 1969 count.[12] Some order was brought to citrus taxonomy by two alternative unified classification schemes, those of Chōzaburō Tanaka and Walter Tennyson Swingle, which can be viewed as extreme alternative visions of the same genus.[12][6] Swingle's system, divided the Citrinae subtribe into three groups, the 'primitive citrus' distant relatives, the closer 'near citrus' including citrus-related genera like Atalantia, and the 'true citrus', which included Poncirus, Citrus, Fortunella, Eremocitrus, Microcitrus, and Clymenia, all but the first now viewed to fall within Citrus. His Citrus he likewise subdivided into two subgenera: citrons, pomelos, mandarins, oranges, grapefruits and lemons were placed in subgenus Eucitrus (later called simply subgenus Citrus), while the hardy but slow-growing trees with relatively unpalatable fruit he placed in subgenus Papeda.[13][14][15] His genus Citrus consisted of just 16 species, dividing them further into varieties, and lastly cultivars or hybrids. The Swingle system is generally followed today with much modification; however, there are still large differences in nomenclature between countries and individual scientists.[16] The basis of the 'Tanaka system' (1954) is to provide a separate species name for each cultivar, regardless of whether it is pure or a hybrid of two or more species or varieties, and resulted in 159 identified species.[12] It thus represents an example of taxonomic "splitting",[17] and in assigning separate species names to horticultural variants does not conform to the standard species concept.[12] Tanaka also divides into subgenera, but differently than Swingle, introducing Archicitrus (which he subdivided into five sections, Papeda, Limonellus, Aruntium, Citrophorum and Cephalocitrus) and Metacitrus (divided into Osmocitrus, Acrumen and Pseudo fortunella).[14][18] A 1969 analysis by Hodgson intended to harmonize the two schemes accepting 36 species.[12][15] These initial attempts at Citrus systematization all predate the recognition, which began to gain traction in the mid-1970s, that the majority of cultivars represent hybrids of just three species, citron, mandarin and pomelo.[18][15][19]
Phylogenetic analysis confirms the hybrid origin of most citrus cultivars, indicating a small number of founder species. While the subgenera suggested by Tanaka proved similar to the phylogenetic divisions, Swingle's subgenera were polyphyletic,[18] and hence do not represent valid taxonomy. Historical genera are also dubious. Kumquats had been placed in a separate genus Fortunella, while two genera were suggested by Swingle for the Australian limes, Microcitrus and Eremocitrus. However, genomic analysis has pointed to these groups nesting within the phylogenetic tree of the other citrus plants. Since their placement in distinct genera would make Citrus a polyphyletic grouping, it has been suggested that all of these are correctly members of a single genus, Citrus.[6][20] There remains a lack of consensus as to which wild plants and hybrids merit distinct species status, a phenomenon exacerbated by the prior failure to correctly identify the genetically pure citrus strains and distinguish them from hybrids.[6]
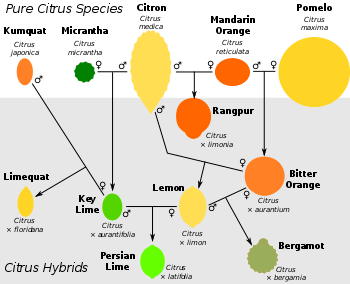
Core species and hybrids
Most commercial varieties are descended from one or more of the 'core species', citrons, mandarins, and pomelos, which share in common a complex floral anatomy that gives rise to more complex fruit. These core species, and to a lesser extent other citrus, have given rise to a wide variety of hybrids for which the naming is inconsistent. The same common names may be given to different species, citrus hybrids or mutations. For example, citrus with green fruit tend to be called 'limes' independent of their origin: Australian limes, Musk limes, Key limes, kaffir limes, Rangpur limes, sweet limes and wild limes are all genetically distinct. Fruit with similar ancestry may be quite different in name and traits (e.g. grapefruit, common oranges, and ponkans, all pomelo-mandarin hybrids). Many traditional citrus groups, such as true sweet oranges and lemons, seem to be bud sports, clonal families of cultivars that have arisen from distinct spontaneous mutations of a single hybrid ancestor.[9][6]
Ancestral species
.jpg)
Mandarins
Mandarin oranges (tangerines, satsumas - Citrus reticulata) are one of the basic species, but the name mandarin is also used more generally for all small, easily peeled citrus, including a large range of hybrids.[21] Swingle saw three species of mandarin, while Tanaka identified five groups with a total of 36 species. Webber (1948) divided them into four groups, king, satsuma, mandarin, and tangerine, and Hodgson (1967) saw in them four species.[6][21] Genomic analysis suggests just one species, Citrus reticulata, with even the divergent Tachibana mandarin (Tanaka's Citrus tachibana), native to Taiwan, the Ryukyu Islands and southern Japan, showing enough genomic similarity with mandarins of the East-Asian mainland to be considered just a subspecies.[6]
Many varieties commercially called mandarins are actually hybrids.[3][22] Wu, et al., divided mandarins into three types, based on their degree of hybridization. In addition to genetically pure mandarins, a second type are the result of hybridization with pomelos followed by subsequent backcrossing with mandarins to retain only a few pomelo traits. The third type arose more recently from the crossing of these hybrids again with pomelos or sweet oranges (which are themselves crosses of hybrid mandarins and pomelos). This produces mandarins with more, longer stretches of pomelo DNA.[6] Some commercial mandarins are hybrids with lemons, while several were found to have a significant contribution (35-65%) from papedas.[21]
The mangshanyegan (C. mangshanensis), known colloquially as the 'mangshan mandarin', is a genetically distinct citrus species.[6]
Pomelos

The pomelo (Citrus maxima), a second of the core species from which most citrus hybrids have derived, is native to southeast Asia. Among the hybrids deriving from mandarin/pomelo crosses, there is a direct correlation between the proportion of pomelo DNA in the hybrid and fruit size, while the more palatable mandarins are those that have received specific genes from pomelos that alter their acidity.[6] Some of the more common pomelos are genetically pure, while a number have a single small region of introgressed mandarin DNA on one chromosome, the result of a cross followed by extensive backcrossing with pomelo.[23]
Citrons
Varieties of true (non-hybrid) Citron (Citrus medica) have distinctly different forms. The citron usually propagates by cleistogamy, a self-pollination within an unopened flower, and this results in the lowest levels of heterozygosity among the citrus species.[6] Because of this, it will generally serve as the male parent of any hybrid progeny. Many citron varieties were proven to be non-hybrids despite their rather dramatic morphological differences;[18][24][25][5][26][27] however, the florentine citron is probably of hybrid origin. Genetic analysis of citrons has shown that they divide into three groups. One cluster consists of wild citrons that originated in China and produce non-fingered fruit with pulp and seeds. A second cluster, also native to China, consist of the fingered citrons, most of which are seedless and must be propagated artificially. The third cluster represents the Mediterranean citrons, thought to have originally been introduced there from India.[28]
Some fingered citron varieties are used in buddhist offerings, and some more common varieties are used as the etrog in the Jewish harvest festival of Sukkot. There is also a specific variety of citron called etrog. The Mountain citron is a complex citrus hybrid that only includes trace amounts of true citron.[21]
Kumquats
Kumquats are a separate species, with few hybrids. Carl Peter Thunberg originally classified the kumquats as Citrus japonica in his 1784 book, Flora Japonica. In 1915, Swingle reclassified them in a separate genus, Fortunella, named in honor of Robert Fortune. Seven species of Fortunella were generally recognized—F. japonica, F. margarita, F. crassifolia, F. hindsii, F. obovata and F. polyandra, as well as the recently described F. bawangica. Since the kumquat is a cold hardy species, there are many hybrids between common citrus members and the kumquat. Swingle coined a separate hybrid genus for these, which he called × Citrofortunella.
Subsequent study of the many commercial citrus lineages revealed such complexity that the genera could not be separated,[29] and genomic analysis rooted Fortunella within the polyphyletic tree of Citrus.[6][20] Consequently, in accordance with the International Code of Nomenclature for algae, fungi, and plants, the correct genus name reverted to Citrus. The Flora of China unites all kumquats as the single species, Citrus japonica.[30]
Papedas
Swingle coined the Citrus subgenus Papeda to separate its members from the more edible citrus, that also differ from other citrus in that its stamens grow separately, not united at the base.[31] He included in this group the kaffir lime, as well as the Ichang papeda (Citrus ichangensis) and the micrantha (Citrus micrantha). Since the latter two species locate to different branches of the citrus phylogenetic tree, the group would be polyphyletic and not a valid division.[32][18] Papedas have also given rise to hybrids with other citrus.
Australian and New Guinean species

Australian and New Guinean citrus species had been viewed as belonging to separate genera by Swingle, who placed in Microcitrus all but the desert lime, for whcih he used Eremocitrus. However, genomic analysis shows that though they form a distinct clade from other citrus, this is nested within the citrus phylogenetic tree, most closely related to kumquats, suggesting that all these species should be included in the genus Citrus.[18][6] Wu, et al., found that several of the finger lime cultivars were actually hybrids with round lime, and concluded there were just three species among those tested, desert lime (C. glauca), round lime (C. australis) and the finger lime (C. australasica), though their analysis did not include other types previously identified as distinct species.[6] In more limited genomic analysis, the New Guinea wild lime, Clymenia and Oxanthera (false orange) all cluster with the Australian limes as members of Citrus.[20][33] The outback lime is a desert lime agriculturally-selected for more commercial traits, while Some commercial varieties of the Australian lime are hybrids with mandarins, lemons, and/or sweet oranges, and bear . Clymenia, will hybridize with kumquats and some limes.
Hybrids
Citrus hybrids include many varieties and species that have been selected by plant breeders. This is done not only for the useful characteristics of the fruit, but also for plant size and growth characteristics such as cold-tolerance. Some citrus hybrids occurred naturally, and others have been deliberately created, either by cross pollination and selection among the progeny, or (rarely, and only recently) as somatic hybrids. The aim of plant breeding of hybrids is to use two or more different citrus varieties or species, in order to get traits intermediate between those of the parents, or to transfer individual desirable traits of one parent into the other. In some cases, particularly with the natural hybrids, it has been viewed as hybrid speciation and the new plants have been viewed as different species from any of their parents. Citrus hybrids have often been given unique hybrid names, marked with a multiplication sign after the word "Citrus", for example the Key lime is Citrus × aurantifolia, and also are referred to by joining the names of the crossed species or hybrids that produced them, as with sunquat – Citrus limon × japonica. Styling a hybrid as such a cross between two species can present challenges. In some cases the parental species that gave rise to a hybrid have yet to be determined, while genotyping reveals some hybrids to descend from three or more ancestral species.
Labelling of hybrids
Hybrid taxonomy is inconsistent. There is disagreement over whether to assign species names to hybrids, and even modern hybrids of known parentage are sold under general names that give little information about their ancestry, or even give technically incorrect information. This can be a problem for those who can eat only some citrus varieties. Drug interactions with chemicals found in some citrus, including grapefruit and Seville oranges,[34][35] make the ancestry of citrus fruit of interest; many commonly sold citrus varieties are grapefruit hybrids[36][37] or pomelo-descended grapefruit relatives. One medical review has advised patients on medication to avoid all citrus juice,[34] although some citrus fruits contain no furanocoumarins.[37] Citrus allergies can also be specific to only some fruit or some parts of some fruit.[38][39][40]
Major citrus hybrids


The most common citrus hybrids that are sometimes treated as a species by themselves, especially in folk taxonomy, are:
- Orange: a name used for several distinct crosses between pomelo and a mandarin orange. They have the orange color of the mandarin in their outer peels and segments, and are easier to peel than the pomelos. Oranges are all intermediate between the two ancestors in size, flavor and shape.[3][41] The sour orange and sweet orange both arose from mandarin-pomelo crosses, the former with a pure mandarin, the latter with a mandarin already containing small amounts of pomelo.[6]
- Grapefruit: Grapefruits are more akin to the pomelo ancestor,[42] resulting from a natural cross between a sweet orange and a pomelo.[23] The 'cocktail grapefruit', or Mandelo, is distinct, instead the product of a low-acid pomelo variety hybridized with a mandarin that itself was a cross between two distinct mandarin stocks.[6]
- Lemon: Most lemons are from one common ancestor, and diverged by mutation. The ancestor was a hybrid between a male citron and a female sour orange, itself a pomelo/mandarin hybrid; citrons contribute half of the genome, while the other half is divided between pomelo and mandarin.[6][43] There are other hybrids also known as 'lemons'. Rough lemons arose from a cross between citron and mandarin, without the pomelo contribution found in 'true' lemons, while the Meyer lemon comes from a citron crossed with a sweet (as opposed to sour) orange.[8]
- Limes: A highly diverse group of hybrids go by this name. Rangpur limes, like rough lemons, arose from crosses between citron and mandarin. The sweet limes, so-called due to their low acid pulp and juice, come from crosses of citron with either sweet or sour oranges, while the Key lime arose from a cross between a citron and a micrantha.[8]
All of these hybrids have in turn been bred back with their parent stocks or with other citrus to form a broader array of fruits, for example the Ponderosa lemon (Citrus limon × medica) and Florentine citron (Citrus x limonimedica) are both lemon/citron hybrids, and Oroblanco is a grapefruit/pomelo mix, while tangelos are mandarin/pomelo or mandarin/grapefruit hybrids, orangelos result from grapefruit backcrossed with sweet orange, and a sweet orange backcrossed with a tangerine gives the tangor.[23] One lumia, a member of the sweet lemons, is the product of crossing a lemon with a pomelo/citron hybrid, though another lumia variety, the Pomme d'Adam, is a micrantha/citron cross, like the Key lime. The most common and commercially popular 'limes' are the Persian limes, Key lime/lemon hybrids that combine the genetic lineages of four ancestral citrus species: mandarin, pomelo, citron and micrantha.[8]
While most other citrus are diploid, many of the Key lime hybrid progeny have unusual chromosome numbers. For example, the Persian lime is triploid, deriving from a diploid Key lime gamete and a haploid lemon ovule. A second group of Key lime hybrids, including the Tanepao lime and Madagascar lemon, are also triploid but instead seem to have arisen from a backcross of a diploid Key lime ovule with a citron haploid gamete. The 'Giant Key lime' owes its increased size to a spontaneous duplication of the entire diploid Key lime genome to produce a tetraploid.[8]
Hybrids from other citrus species
While most citrus hybrids derive from the three core species, hybrids have also been derived from the micrantha, the Ichang papeda and the kumquat. The best known hybrid from micrantha is the Key lime (or Mexican lime), derived from the breeding of a male citron and a female micrantha. Several citrus varieties are Ichang papeda/mandarin crosses (for which Swingle coined the term ichandarin[19]), including Sudachi and Yuzu (which also includes smaller contributions from pomelo and kumquat).[21] Other more exotic citrus have likewise proved hybrids that include papeda. For example, the Indian wild orange, once suggested as a possible ancestor of today's cultivated citrus fruits,[44] yielded conflicting phylogenetic placements in more limited genetic analysis,[18][45] but study of nuclear markers and chloroplast DNA showed it to be of maternal citron lineage, with further genetic contributions from mandarin and papeda.[21] The wild Mountain citron, native to the Malay peninsula and called Citrus halimii at the time of its discovery,[12] has been found to be a complex mix, being half papeda (type not specified), the other half including kumquat, mandarin and pomelo, with only trace amounts of citron.[21]
Citrofortunella
A large group of commercial hybrids involve the kumquat, Fortunella in the Swingle system. Citrofortunella was coined as a genus containing intergeneric hybrids between members of the Citrus and the Fortunella, and is named after its parent genera.[46] Such hybrids often combine the cold hardiness of the Fortunella with some edibility properties of the Citrus species. As members of a hybrid genus, Citrofortunellas are marked with the multiplication sign before the genus name, for example × Citrofortunella microcarpa. With the placement of kumquats within Citrus, Citrofortunella are no longer viewed as being intergeneric hybrids. All likewise belong in Citrus, while Citrofortunella as a distinct genus name would no longer be valid.[29] Examples of the Citrofortunella include the calamondin, limequat and yuzuquat, crossing kumquat with tangerine, Key lime and yuzu respectively.
Chimera
Graft-chimaeras, also called graft hybrids, can occur in Citrus. The cells are not somatically fused but rather mix the tissues from scion and rootstock after grafting, a popular example the Bizzaria orange. In formal usage, these are marked with a plus sign "+" instead with an "x".
Citrus relatives
Trifoliate orange

The trifoliate orange is a cold hardy plant distinguishable by its compound leaves with three leaflets and their deciduous nature, but is close enough to the genus Citrus to be used as a rootstock.[47] It was long thought to be the only member of the Poncirus genus, until a second species, Poncirus polyandra, was discovered in Yunnan (China) in the 1980s.[32] Swingle placed the trifoliate orange in its own genus, Poncirus. Early phylogenetic analysis nested Poncirus within the citrus, suggesting they all belong to a single genus,[18][48][20][32] but the genomic sequencing of Wu, et al., placed it outside the cluster representing Citrus, and the authors retained Poncirus as a separate genus.[6] As with kumquats, the trifolate orange does not naturally interbreed with core taxa due to different flowering times,[11] but hybrids have been produced artificially.
Citroncirus
Citroncirus was coined in the Swingle system as a hybrid genus, resulting from crosses between members of the genus Citrus and the closely related Poncirus.[46] As a hybrid genus, it can be represented as "× Citroncirus". The group includes the citrange, a hybrid between the trifoliate and sweet oranges, and the citrumelo, a hybrid of trifolate orange and 'Duncan' grapefruit. Were Poncirus to be subsumed into Citrus, these hybrids would no longer be intergeneric, and would all likewise fall within Citrus.
Wild lime
The wild lime—native to southern Florida and Texas in the United States, Mexico, Central America, the Caribbean, and South America as far south as Paraguay—does not belong in the genus Citrus. It is part of another genus in the family Rutaceae, Zanthoxylum (which also includes Sichuan pepper), and was classified as Zanthoxylum fagara by Charles Sprague Sargent.[49]
See also
References
- 1 2 G. A. Moore (Sep 2001). "Oranges and lemons: clues to the taxonomy of Citrus from molecular markers". Trends in Genetics. 17: 536–40. doi:10.1016/s0168-9525(01)02442-8. PMID 11525837.
- ↑ Velasco, Riccardo; Licciardello, Concetta (2014). "A genealogy of the citrus family". Nature Biotechnology. 32: 640–642.
- 1 2 3 4 G Albert Wu; et al. "Sequencing of diverse mandarin, pomelo and orange genomes reveals complex history of admixture during citrus domestication". Nature Biotechnology. 32: 656–662. doi:10.1038/nbt.2906. PMC 4113729. PMID 24908277.
- ↑ Li, Xiaomeng; Xie, Rangjin; Lu, Zhenhua; Zhou, Zhiqin (2010). "The Origin of Cultivated Citrus as Inferred from Internal Transcribed Spacer and Chloroplast DNA Sequence and Amplified Fragment Length Polymorphism Fingerprints". Journal of the American Society for Horticultural Science. 135: 341–350.
- 1 2 Nicolosi, Elisabetta; La Malfi, Stefano; El-Otmani, Mohamed; Neghi, Moshe; Goldschmidt, Eliezer E (2005). "The Search for the Authentic Citron (Citrus medicus L.): Historic and Genetic Analysis". HortScience. 40: 1963–1968.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 Wu, Guohong Albert; Terol, Javier; Ibanez, Victoria; López-García, Antonio; Pérez-Román, Estela; Borredá, Carles; Domingo, Concha; Tadeo, Francisco R; Carbonell-Caballero, Jose; Alonso, Roberto; Curk, Franck; Du, Dongliang; Ollitrault, Patrick; Roose, Mikeal L. Roose; Dopazo, Joaquin; Gmitter Jr, Frederick G.; Rokhsar, Daniel; Talon, Manuel (2018). "Genomics of the origin and evolution of Citrus". Nature. 554: 311–316. doi:10.1038/nature25447. and Supplement
- ↑ Andrés García Lor (2013). Organización de la diversidad genética de los cítricos (PDF) (Thesis). p. 79. (in Spanish)
- 1 2 3 4 5 Curk, Franck; Ollitrault, Frédérique; Garcia-Lor, Andres; Luro, François; Navarro, Luis; Ollitrault, Patrick (2016). "Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers". Annals of Botany. 11: 565–583. doi:10.1093/aob/mcw005. PMC 4817432.
- 1 2 Curk, Franck; Ancillo, Gema; Garcia-Lor, Andres; Luro, François; Perrier, Xavier; Jacquemoud-Collet, Jean-Pierre; Navarro, Luis; Ollitrault, Patrick (2014). "Next generation haplotyping to decipher nuclear genomic interspecific admixture in Citrus species: analysis of chromosome 2". BMC Genetics. 15: 152.
- ↑ Penjor, Tshering; Yamamoto, Masashi; Uehara, Miki; Ide, Manami; Matsumoto, Natsumi; Matsumoto, Ryoji; Nagano, Yukio (2013). "Phylogenetic Relationships of Citrus and Its Relatives Based on matK Gene Sequences". PLOS One. 8: e62574. doi:10.1371/journal.pone.0062574.
- 1 2 Froelicher, Yann; Mouhaya, Wafa; Bassene, Jean-Baptiste; Costantino, Gilles; Kamiri, Mourad; Luro, Francois; Morillo, Raphael; Ollitrault, Patrick (2011). "New universal mitochondrial PCR markers reveal new information on maternal citrus phylogeny". Tree Genetics & Genomes. 7: 49–61. doi:10.1007/s11295-010-0314-x.
- 1 2 3 4 5 6 Stone, Benjamin C; Lowry, J Brian; Scora, R W; Jong, Kwiton (1973). "Citrus halimii: A New Species from Malaya and Peninsular Thailand". Biotropica. 5: 102–110. doi:10.2307/2989659.
- ↑ Wang, Xia; et al. (2017). "Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction". Nature Genetics. 49: 765–772. doi:10.1038/ng.3839.
- 1 2 Sharma, Girish; Sharma, O. C.; Thakur, B. S. (2009). Systematics of Fruit Crops. New India Publishing Agency. pp. 91–92.
- 1 2 3 Nicolosi, Elisabetta (2007), Kahn, Iqrqr Ahmad, ed., "Origin and Taxonomy", Citrus Genetics, Breeding and Biotechnology, CAB International, pp. 19–43
- ↑ Krueger, R.R.; Navarro, L. (2007), Kahn, Iqrqr Ahmad, ed., "Citrus Germplasm Resources", Citrus Genetics, Breeding and Biotechnology, CAB International, p. 49
- ↑ Page, Martin (2008). Growing Citrus: The Essential Gardener's Guide. Timber Press. p. 30. ISBN 978-0-88192-906-5.
- 1 2 3 4 5 6 7 8 Nicolosi, E; Deng, Z. N.; Gentile, A.; La Malfa, S.; Continella, G.; Tribulato, E. (2000). "Citrus phylogeny and genetic origin of important species as investigated by molecular markers". Theoretical and Applied Genetics. 100: 1155–1166.
- 1 2 Shimizu, Tokurou; Kitajima, Akira; Nonaka, Keisuke; Yoshioka, Terutaka; Ohta, Satoshi; Goto, Shingo; Toyoda, Atsushi; Fujiyama, Asao; Mochizuki, Takako; Nagasaki, Hideki; Kaminuma, Eli; Nakamura, Yasukazu (2016). "Hybrid Origins of Citrus Varieties Inferred from DNA Marker Analysis of Nuclear and Organelle Genomes". PLOS One. 11: e0166969. doi:10.1371/journal.pone.0166969.
- 1 2 3 4 Bayer, Randall J; Mabberly, David J; Morton, Cynthia; Miller, Cathy H.; Sharma, Ish K; Pfiel, Bernard E; Rich, Sarah; Hitchcock, Roberta; Sykes, Steve (2009). "A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences". American Journal of Botany. 96 (3): 668–685. doi:10.3732/ajb.0800341.
- 1 2 3 4 5 6 7 Garcia-Lor, Andres; Luro, François; Ollitrault, Patrick; Navarro, Luis (2015). "Genetic diversity and population structure analysis of mandarin germplasm by nuclear, chloroplastic and mitochondrial markers". Tree Genetics & Genomes. 11: e123. doi:10.1007/s11295-015-0951-1.
- ↑ de Oliveira, Roberto Pedroso; Radmann, Elizete Beatriz (2005). "Genetic similarity of citrus fresh fruit market cultivars". Revista Brasileira de Fruticultura. 27: 332–334. doi:10.1590/S0100-29452005000200037.
- 1 2 3 Oueslati, Amel; Salhi-Hannachi, Amel; Luro, François; Vignes, Hélène; Mournet, Pierre; Ollitrault, Patrick (2017). "Genotyping by sequencing reveals the interspecific C. maxima / C. reticulata admixture along the genomes of modern citrus varieties of mandarins, tangors, tangelos, orangelos and grapefruits". PLOS One. 12: e0185618. doi:10.1371/journal.pone.0185618.
- ↑ Barkley, Noelle A.; Roose, Mikeal L.; Krueger, Robert R.; Federici, Claire T. (2006). "Assessing genetic diversity and population structure in a Citrus germplasm collection utilizing simple sequence repeat markers (SSRs)". Theoretical and Applied Genetics. 112: 1519–1531.
- ↑ Abkenar, Asad Asadi; Isshiki, Shiro; Tashiro, Yosuke (2004). "Phylogenetic relationships in the "true citrus fruit trees" revealed by PCR-RFLP analysis of cpDNA". Scientia Horticulturae. 102: 233–242.
- ↑ King, C. A. (1943). "Chromosome Numbers in the Subfamily Aurantioideae with Special Reference to the Genus Citrus". Botanical Gazette. 104: 602–611. JSTOR 2472147.
- ↑ Carvalhoa, R.; Soares Filhob, W.S.; Brasileiro-Vidala, A.C.; Guerra, M. (2005). "The relationships among lemons, limes and citron: a chromosomal comparison". Cytogenetic and Genome Research. 109: 276–282.
- ↑ Ramadugu, Chandrika; Keremane, Manjunath L; Hu, Xulan; Karp, David; Frederici, Claire T; Kahn, Tracy; Roose, Mikeal L; Lee, Richard F. (2015). "Genetic analysis of citron (Citrus medica L.) using simple sequence repeats and single nucleotide polymorphisms". Scientia Horticulturae. 195: 124–137.
- 1 2 Mabberley, D. J. (2004). "Citrus (Rutaceae): A Review of Recent Advances in Etymology, Systematics and Medical Applications". Blumea. 49: 481–498.
- ↑ Zhang, Dianxiang; Hartley, Thomas G.; Mabberley, David J. (2008), "Rutaceae", Flora of China, 11, pp. 51–97
- ↑ Mabberley, David (1997). "A classification for edible Citrus (Rutaceae)" (PDF). Telopea. 7: 167–172.
- 1 2 3 Garcia-Lor, Andres; Curk, Franck; Snoussi-Trifa, Hager; Morillon, Raphael; Ancillo, Gema; Luro, François; Navarro, Luis; Ollitrault, Patrick (2011). "A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the 'true citrus fruit trees' group (Citrinae, Rutaceae) and the origin of cultivated species". Annals of Botany. 111: 1–19. doi:10.1093/aob/mcs227. PMC 3523644.
- ↑ Oueslati, Amel; Ollitrault, Frederique; Baraket, Ghada; Salhi-Hannachi, Amel; Navarro, Luis; Ollitrault, Patrick (2016). "Towards a molecular taxonomic key of the Aurantioideae subfamily using chloroplastic SNP diagnostic markers of the main clades genotyped by competitive allele-specific PCR". BMC Genetics. 17: 118. doi:10.1186/s12863-016-0426-x.
- 1 2 Saito, M; Hirata-Koizumi, M; Matsumoto, M; Urano, T; Hasegawa, R (2005). "Undesirable effects of citrus juice on the pharmacokinetics of drugs: focus on recent studies". Drug Safety. 28 (8): 677–94. doi:10.2165/00002018-200528080-00003. PMID 16048354.
- ↑ Bailey, David G. (2010). "Fruit juice inhibition of uptake transport: a new type of food-drug interaction". British Journal of Clinical Pharmacology. 70 (5): 645–55. doi:10.1111/j.1365-2125.2010.03722.x. PMC 2997304. PMID 21039758.
- ↑ Morton, Julia F. (1987). "Tangelo". Fruits of Warm Climates. Florida Flair Books. pp. 159–160.
- 1 2 Widmer, Wilbur (2006). "One Tangerine/Grapefruit Hybrid (Tangelo) Contains Trace Amounts of Furanocoumarins at a Level Too Low To Be Associated with Grapefruit/Drug Interactions". Journal of Food Science. 70 (6): c419–22. doi:10.1111/j.1365-2621.2005.tb11440.x.
- ↑ Bourrier, T; Pereira, C (2013). "Allergy to citrus juice". Clinical and Translational Allergy. 3 (Suppl 3): P153. doi:10.1186/2045-7022-3-S3-P153. PMC 3723546.
- ↑ Cardullo, AC; Ruszkowski, AM; DeLeo, VA (1989). "Allergic contact dermatitis resulting from sensitivity to citrus peel, geraniol, and citral". Journal of the American Academy of Dermatology. 21 (2 Pt 2): 395–7. doi:10.1016/s0190-9622(89)80043-x. PMID 2526827.
- ↑ Boonpiyathad, S (2013). "Chronic angioedema caused by navel orange but not citrus allergy: case report". Clinical and Translational Allergy. 3 (Suppl 3): P159. doi:10.1186/2045-7022-3-S3-P159. PMC 3723846.
- ↑ Xu, Qiang; et al. (2013). "The draft genome of sweet orange (Citrus sinensis)". Nature Genetics. 45: 59–66.
- ↑ Corazza-Nunes, M.J. "Assessment of genetic variability in grapefruits (Citrus paradisi Macf.) and pomelos (C. maxima (Burm.) Merr.) using RAPD and SSR markers". Euphytica. 126: 169–176. doi:10.1023/A:1016332030738.
- ↑ Gulson, O.; Roose, M. L. (2001). "Lemons: Diversity and Relationships with Selected Citrus Genotypes as Measured with Nuclear Genome Markers" (PDF). Journal of the American Society for Horticultural Science. 126: 309–317.
- ↑ Malik, S. K.; Chaudhury, R.; Dhariwal, O. P.; kalia, R. K. (2006). "Collection and characterization of Citrus indica Tanaka and C. macroptera Montr.: wild endangered species of northeastern India". Genetic Resources and Crop Evolution. 53: 1485–1493.
- ↑ Fang, Dequi; Krueger, Robert R; Roose, Mikeal L (1998). "Phylogenetic Relationships among Selected Citrus Germplasm Accessions Revealed by Inter-simple Sequence Repeat Markers" (PDF). Journal of the American Society for Horticultural Science. 123: 612–617.
- 1 2 Ingram, J.; Moore, H. e. (1976). "Rutaceae". Baileya. 19: 169–171.
- ↑ Kaneyoshi, J.; Kobayashi, S. (2000). "Genetic Transformation of Poncirus trifoliata (Trifoliate Orange)". Transgenic Trees. Biotechnology in Agriculture and Forestry. 44. pp. 212–220.
- ↑ de Araújo, Edson Freitas; Queiroza, Luciano Paganuccide; Machado, Marcos Antônio (2003). "What is Citrus? Taxonomic implications from a study of cp-DNA evolution in the tribe Citreae (Rutaceae subfamily Aurantioideae)". 1: 55–62. doi:10.1078/1439-6092-00058.
- ↑ "Zanthoxylum fagara". Germplasm Resources Information Network (GRIN). Agricultural Research Service (ARS), United States Department of Agriculture (USDA). Retrieved 2009-11-27.
External links
| Wikimedia Commons has media related to |
- USDA Citrus Genome Database
- USDA Classification
- Citrus Pages A comprehensive article on citrus taxonomy
- Fruits of warm climates
- International Code of Botanical Nomenclature (Tokyo Code)
- GRIN database for Species of Citrus
.jpg)