Organism
In biology, an organism (from Greek: ὀργανισμός, organismos) is any individual entity that embodies the properties of life. It is a synonym for "life form".
_01.jpg)

Organisms are classified by taxonomy into groups such as multicellular animals, plants, and fungi; or unicellular microorganisms such as protists, bacteria, and archaea.[1] All types of organisms are capable of reproduction, growth and development, maintenance, and some degree of response to stimuli. Humans, squids, mushrooms, and vascular plants are examples of multicellular organisms that differentiate specialized tissues and organs during development.
An organism may be either a prokaryote or a eukaryote. Prokaryotes are represented by two separate domains – bacteria and archaea. Eukaryotic organisms are characterized by the presence of a membrane-bound cell nucleus and contain additional membrane-bound compartments called organelles (such as mitochondria in animals and plants and plastids in plants and algae, all generally considered to be derived from endosymbiotic bacteria).[2] Fungi, animals and plants are examples of kingdoms of organisms within the eukaryotes.
Estimates on the number of Earth's current species range from 2 million to 1 trillion,[3] of which over 1.7 million have been documented.[4] More than 99% of all species, amounting to over five billion species,[5] that ever lived are estimated to be extinct.[6][7]
In 2016, a set of 355 genes from the last universal common ancestor (LUCA) of all organisms was identified.[8][9]
Etymology
The term "organism" (from Greek ὀργανισμός, organismos, from ὄργανον, organon, i.e. "instrument, implement, tool, organ of sense or apprehension")[10][11] first appeared in the English language in 1703 and took on its current definition by 1834 (Oxford English Dictionary). It is directly related to the term "organization". There is a long tradition of defining organisms as self-organizing beings, going back at least to Immanuel Kant's 1790 Critique of Judgment.[12]
Definitions
An organism may be defined as an assembly of molecules functioning as a more or less stable whole that exhibits the properties of life. Dictionary definitions can be broad, using phrases such as "any living structure, such as a plant, animal, fungus or bacterium, capable of growth and reproduction".[13] Many definitions exclude viruses and possible man-made non-organic life forms, as viruses are dependent on the biochemical machinery of a host cell for reproduction.[14] A superorganism is an organism consisting of many individuals working together as a single functional or social unit.[15]
There has been controversy about the best way to define the organism[16][17][18][19][20][21][22][23][24] and indeed about whether or not such a definition is necessary.[25][26] Several contributions[27] are responses to the suggestion that the category of "organism" may well not be adequate in biology.[28]
Viruses
Viruses are not typically considered to be organisms because they are incapable of autonomous reproduction, growth or metabolism. Although some organisms are also incapable of independent survival and live as obligatory intracellular parasites, they are capable of independent metabolism and procreation. Although viruses have a few enzymes and molecules characteristic of living organisms, they have no metabolism of their own; they cannot synthesize and organize the organic compounds from which they are formed. Naturally, this rules out autonomous reproduction: they can only be passively replicated by the machinery of the host cell. In this sense, they are similar to inanimate matter.
While viruses sustain no independent metabolism and thus are usually not classified as organisms, they do have their own genes, and they do evolve by mechanisms similar to the evolutionary mechanisms of organisms. Thus, an argument that viruses should be classed as living organisms is their ability to undergo evolution and replicate through self-assembly. However, some scientists argue that viruses neither evolve nor self-reproduce. Instead, viruses are evolved by their host cells, meaning that there was co-evolution of viruses and host cells. If host cells did not exist, viral evolution would be impossible. This is not true for cells. If viruses did not exist, the direction of cellular evolution could be different, but cells would nevertheless be able to evolve. As for the reproduction, viruses totally rely on hosts' machinery to replicate.[29] The discovery of viruses with genes coding for energy metabolism and protein synthesis fuelled the debate about whether viruses are living organisms. The presence of these genes suggested that viruses were once able to metabolize. However, it was found later that the genes coding for energy and protein metabolism have a cellular origin. Most likely, these genes were acquired through horizontal gene transfer from viral hosts.[29]
Chemistry
Organisms are complex chemical systems, organized in ways that promote reproduction and some measure of sustainability or survival. The same laws that govern non-living chemistry govern the chemical processes of life. It is generally the phenomena of entire organisms that determine their fitness to an environment and therefore the survivability of their DNA-based genes.
Organisms clearly owe their origin, metabolism, and many other internal functions to chemical phenomena, especially the chemistry of large organic molecules. Organisms are complex systems of chemical compounds that, through interaction and environment, play a wide variety of roles.
Organisms are semi-closed chemical systems. Although they are individual units of life (as the definition requires), they are not closed to the environment around them. To operate they constantly take in and release energy. Autotrophs produce usable energy (in the form of organic compounds) using light from the sun or inorganic compounds while heterotrophs take in organic compounds from the environment.
The primary chemical element in these compounds is carbon. The chemical properties of this element such as its great affinity for bonding with other small atoms, including other carbon atoms, and its small size making it capable of forming multiple bonds, make it ideal as the basis of organic life. It is able to form small three-atom compounds (such as carbon dioxide), as well as large chains of many thousands of atoms that can store data (nucleic acids), hold cells together, and transmit information (protein).
Macromolecules
Compounds that make up organisms may be divided into macromolecules and other, smaller molecules. The four groups of macromolecule are nucleic acids, proteins, carbohydrates and lipids. Nucleic acids (specifically deoxyribonucleic acid, or DNA) store genetic data as a sequence of nucleotides. The particular sequence of the four different types of nucleotides (adenine, cytosine, guanine, and thymine) dictate many characteristics that constitute the organism. The sequence is divided up into codons, each of which is a particular sequence of three nucleotides and corresponds to a particular amino acid. Thus a sequence of DNA codes for a particular protein that, due to the chemical properties of the amino acids it is made from, folds in a particular manner and so performs a particular function.
These protein functions have been recognized:
- Enzymes, which catalyze all of the reactions of metabolism
- Structural proteins, such as tubulin, or collagen
- Regulatory proteins, such as transcription factors or cyclins that regulate the cell cycle
- Signaling molecules or their receptors such as some hormones and their receptors
- Defensive proteins, which can include everything from antibodies of the immune system, to toxins (e.g., dendrotoxins of snakes), to proteins that include unusual amino acids like canavanine
A bilayer of phospholipids makes up the membrane of cells that constitutes a barrier, containing everything within the cell and preventing compounds from freely passing into, and out of, the cell. Due to the selective permeability of the phospholipid membrane, only specific compounds can pass through it.
Structure
All organisms consist of structural units called cells; some contain a single cell (unicellular) and others contain many units (multicellular). Multicellular organisms are able to specialize cells to perform specific functions. A group of such cells is a tissue, and in animals these occur as four basic types, namely epithelium, nervous tissue, muscle tissue, and connective tissue. Several types of tissue work together in the form of an organ to produce a particular function (such as the pumping of the blood by the heart, or as a barrier to the environment as the skin). This pattern continues to a higher level with several organs functioning as an organ system such as the reproductive system, and digestive system. Many multicellular organisms consist of several organ systems, which coordinate to allow for life.
Cell
The cell theory, first developed in 1839 by Schleiden and Schwann, states that all organisms are composed of one or more cells; all cells come from preexisting cells; and cells contain the hereditary information necessary for regulating cell functions and for transmitting information to the next generation of cells.
There are two types of cells, eukaryotic and prokaryotic. Prokaryotic cells are usually singletons, while eukaryotic cells are usually found in multicellular organisms. Prokaryotic cells lack a nuclear membrane so DNA is unbound within the cell; eukaryotic cells have nuclear membranes.
All cells, whether prokaryotic or eukaryotic, have a membrane, which envelops the cell, separates its interior from its environment, regulates what moves in and out, and maintains the electric potential of the cell. Inside the membrane, a salty cytoplasm takes up most of the cell volume. All cells possess DNA, the hereditary material of genes, and RNA, containing the information necessary to build various proteins such as enzymes, the cell's primary machinery. There are also other kinds of biomolecules in cells.
All cells share several similar characteristics of:[30]
- Reproduction by cell division (binary fission, mitosis or meiosis).
- Use of enzymes and other proteins coded by DNA genes and made via messenger RNA intermediates and ribosomes.
- Metabolism, including taking in raw materials, building cell components, converting energy, molecules and releasing by-products. The functioning of a cell depends upon its ability to extract and use chemical energy stored in organic molecules. This energy is derived from metabolic pathways.
- Response to external and internal stimuli such as changes in temperature, pH or nutrient levels.
- Cell contents are contained within a cell surface membrane that contains proteins and a lipid bilayer.
Evolution
Last universal common ancestor

The last universal common ancestor (LUCA) is the most recent organism from which all organisms now living on Earth descend.[31] Thus it is the most recent common ancestor of all current life on Earth. The LUCA is estimated to have lived some 3.5 to 3.8 billion years ago (sometime in the Paleoarchean era).[32][33] The earliest evidence for life on Earth is graphite found to be biogenic in 3.7 billion-year-old metasedimentary rocks discovered in Western Greenland[34] and microbial mat fossils found in 3.48 billion-year-old sandstone discovered in Western Australia.[35][36] Although more than 99 percent of all species that ever lived on the planet are estimated to be extinct,[6][7] there are currently 2 million to 1 trillion species of life on Earth.[3]
Information about the early development of life includes input from many different fields, including geology and planetary science. These sciences provide information about the history of the Earth and the changes produced by life. However, a great deal of information about the early Earth has been destroyed by geological processes over the course of time.
All organisms are descended from a common ancestor or ancestral gene pool. Evidence for common descent may be found in traits shared between all living organisms. In Darwin's day, the evidence of shared traits was based solely on visible observation of morphologic similarities, such as the fact that all birds have wings, even those that do not fly.
There is strong evidence from genetics that all organisms have a common ancestor. For example, every living cell makes use of nucleic acids as its genetic material, and uses the same twenty amino acids as the building blocks for proteins. All organisms use the same genetic code (with some extremely rare and minor deviations) to translate nucleic acid sequences into proteins. The universality of these traits strongly suggests common ancestry, because the selection of many of these traits seems arbitrary. Horizontal gene transfer makes it more difficult to study the last universal ancestor.[37] However, the universal use of the same genetic code, same nucleotides, and same amino acids makes the existence of such an ancestor overwhelmingly likely.[38]
Phylogeny
| LUA |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Location of the root
The most commonly accepted location of the root of the tree of life is between a monophyletic domain Bacteria and a clade formed by Archaea and Eukaryota of what is referred to as the "traditional tree of life" based on several molecular studies.[39][40][41][42][43][44] A very small minority of studies have concluded differently, namely that the root is in the domain Bacteria, either in the phylum Firmicutes[45] or that the phylum Chloroflexi is basal to a clade with Archaea and Eukaryotes and the rest of Bacteria as proposed by Thomas Cavalier-Smith.[46]
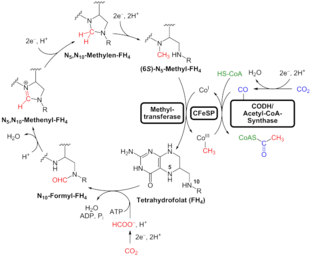
Research published in 2016, by William F. Martin, by genetically analyzing 6.1 million protein-coding genes from sequenced prokaryotic genomes of various phylogenetic trees, identified 355 protein clusters from amongst 286,514 protein clusters that were probably common to the LUCA. The results "depict LUCA as anaerobic, CO2-fixing, H2-dependent with a Wood–Ljungdahl pathway (the reductive acetyl-coenzyme A pathway), N2-fixing and thermophilic. LUCA's biochemistry was replete with FeS clusters and radical reaction mechanisms. Its cofactors reveal dependence upon transition metals, flavins, S-adenosyl methionine, coenzyme A, ferredoxin, molybdopterin, corrins and selenium. Its genetic code required nucleoside modifications and S-adenosylmethionine-dependent methylations." The results depict methanogenic clostria as a basal clade in the 355 lineages examined, and suggest that the LUCA inhabited an anaerobic hydrothermal vent setting in a geochemically active environment rich in H2, CO2, and iron.[47] However, the identification of these genes as being present in LUCA was criticized, suggesting that many of the proteins assumed to be present in LUCA represent later horizontal gene transfers between archaea and bacteria.[48]
Reproduction
Sexual reproduction is widespread among current eukaryotes, and was likely present in the last common ancestor.[49] This is suggested by the finding of a core set of genes for meiosis in the descendants of lineages that diverged early from the eukaryotic evolutionary tree.[50] and Malik et al.[51] It is further supported by evidence that eukaryotes previously regarded as "ancient asexuals", such as Amoeba, were likely sexual in the past, and that most present day asexual amoeboid lineages likely arose recently and independently.[52]
In prokaryotes, natural bacterial transformation involves the transfer of DNA from one bacterium to another and integration of the donor DNA into the recipient chromosome by recombination. Natural bacterial transformation is considered to be a primitive sexual process and occurs in both bacteria and archaea, although it has been studied mainly in bacteria. Transformation is clearly a bacterial adaptation and not an accidental occurrence, because it depends on numerous gene products that specifically interact with each other to enter a state of natural competence to perform this complex process.[53] Transformation is a common mode of DNA transfer among prokaryotes.[54]
Horizontal gene transfer
The ancestry of living organisms has traditionally been reconstructed from morphology, but is increasingly supplemented with phylogenetics – the reconstruction of phylogenies by the comparison of genetic (DNA) sequence.
Sequence comparisons suggest recent horizontal transfer of many genes among diverse species including across the boundaries of phylogenetic "domains". Thus determining the phylogenetic history of a species can not be done conclusively by determining evolutionary trees for single genes.[55]
Biologist Peter Gogarten suggests "the original metaphor of a tree no longer fits the data from recent genome research", therefore "biologists (should) use the metaphor of a mosaic to describe the different histories combined in individual genomes and use (the) metaphor of a net to visualize the rich exchange and cooperative effects of HGT among microbes."[56]
Future of life (cloning and synthetic organisms)
Modern biotechnology is challenging traditional concepts of organism and species. Cloning is the process of creating a new multicellular organism, genetically identical to another, with the potential of creating entirely new species of organisms. Cloning is the subject of much ethical debate.
In 2008, the J. Craig Venter Institute assembled a synthetic bacterial genome, Mycoplasma genitalium, by using recombination in yeast of 25 overlapping DNA fragments in a single step. The use of yeast recombination greatly simplifies the assembly of large DNA molecules from both synthetic and natural fragments.[57] Other companies, such as Synthetic Genomics, have already been formed to take advantage of the many commercial uses of custom designed genomes.
See also
References
- Hine, RS. (2008). A dictionary of biology (6th ed.). Oxford: Oxford University Press. p. 461. ISBN 978-0-19-920462-5.
- Cavalier-Smith T. (1987). "The origin of eukaryotic and archaebacterial cells". Annals of the New York Academy of Sciences. 503 (1): 17–54. Bibcode:1987NYASA.503...17C. doi:10.1111/j.1749-6632.1987.tb40596.x. PMID 3113314.
- Brendan B. Larsen; Elizabeth C. Miller; Matthew K. Rhodes; John J. Wiens (September 2017). "Inordinate Fondness Multiplied and Distributed:The Number of Species on Earth and the New Pie of Life" (PDF). The Quarterly Review of Biology. 92 (3): 230. Retrieved 11 November 2019.
- Anderson, Alyssa M. (2018). "Describing the Undiscovered". Chironomus: Journal of Chironomidae Research (31): 2–3. doi:10.5324/cjcr.v0i31.2887.
- Kunin, W.E.; Gaston, Kevin, eds. (1996). The Biology of Rarity: Causes and consequences of rare – common differences. ISBN 978-0-412-63380-5. Retrieved 26 May 2015.
- Stearns, Beverly Peterson; Stearns, S.C.; Stearns, Stephen C. (2000). Watching, from the Edge of Extinction. Yale University Press. p. preface x. ISBN 978-0-300-08469-6. Retrieved 30 May 2017.
- Novacek, Michael J. (8 November 2014). "Prehistory's Brilliant Future". New York Times. Retrieved 25 December 2014.
- Weiss, Madeline C.; Sousa, Filipa L.; Mrnjavac, Natalia; Neukirchen, Sinje; Roettger, Mayo; Nelson-Sathi, Shijulal; Martin, William F. (2016). "The physiology and habitat of the last universal common ancestor". Nature Microbiology. 1 (9): 16116. doi:10.1038/nmicrobiol.2016.116. PMID 27562259.
- Wade, Nicholas (25 July 2016). "Meet Luca, the Ancestor of All Living Things". New York Times. Retrieved 25 July 2016.
- ὄργανον. Liddell, Henry George; Scott, Robert; A Greek–English Lexicon at the Perseus Project
- "organism". Online Etymology Dictionary.
- Kant I., Critique of Judgment: §64.
- "organism". Chambers 21st Century Dictionary (online ed.). 1999.
- "organism". Oxford English Dictionary (3rd ed.). Oxford University Press. 2004. (Subscription or UK public library membership required.)
- Kelly, Kevin (1994). Out of control: the new biology of machines, social systems and the economic world. Boston: Addison-Wesley. pp. 98. ISBN 978-0-201-48340-6.
- Dupré, J. (2010). "The polygenomic organism". The Sociological Review. 58: 19–99. doi:10.1111/j.1467-954X.2010.01909.x.
- Folse Hj, 3.; Roughgarden, J. (2010). "What is an individual organism? A multilevel selection perspective". The Quarterly Review of Biology. 85 (4): 447–472. doi:10.1086/656905. PMID 21243964.CS1 maint: numeric names: authors list (link)
- Pradeu, T. (2010). "What is an organism? An immunological answer". History and Philosophy of the Life Sciences. 32 (2–3): 247–267. PMID 21162370.
- Gardner, A.; Grafen, A. (2009). "Capturing the superorganism: A formal theory of group adaptation". Journal of Evolutionary Biology. 22 (4): 659–671. doi:10.1111/j.1420-9101.2008.01681.x. PMID 19210588.
- Michod, R E (1999). Darwinian dynamics: evolutionary transitions in fitness and individuality. Princeton University Press. ISBN 978-0-691-05011-9.
- Queller, D.C.; J.E. Strassmann (2009). "Beyond society: the evolution of organismality". Philosophical Transactions of the Royal Society B: Biological Sciences. 364 (1533): 3143–3155. doi:10.1098/rstb.2009.0095. PMC 2781869. PMID 19805423.
- Santelices B. (1999). "How many kinds of individual are there?". Trends in Ecology & Evolution. 14 (4): 152–155. doi:10.1016/s0169-5347(98)01519-5. PMID 10322523.
- Wilson, R (2007). "The biological notion of individual". Stanford Encyclopedia of Philosophy.
- Longo, Giuseppe; Montévil, Maël (2014). Perspectives on Organisms – Springer. Lecture Notes in Morphogenesis. doi:10.1007/978-3-642-35938-5. ISBN 978-3-642-35937-8.
- Pepper, J.W.; M.D. Herron (2008). "Does biology need an organism concept?". Biological Reviews. 83 (4): 621–627. doi:10.1111/j.1469-185X.2008.00057.x. PMID 18947335.
- Wilson, J (2000). "Ontological butchery: organism concepts and biological generalizations". Philosophy of Science. 67: 301–311. doi:10.1086/392827. JSTOR 188676.
- Bateson, P. (2005). "The return of the whole organism". Journal of Biosciences. 30 (1): 31–39. doi:10.1007/BF02705148. PMID 15824439.
- Dawkins, Richard (1982). The Extended Phenotype. Oxford University Press. ISBN 978-0-19-286088-0.
- Moreira, D.; López-García, P.N. (2009). "Ten reasons to exclude viruses from the tree of life". Nature Reviews Microbiology. 7 (4): 306–311. doi:10.1038/nrmicro2108. PMID 19270719.
- The Universal Features of Cells on Earth in Chapter 1 of Molecular Biology of the Cell fourth edition, edited by Bruce Alberts (2002) published by Garland Science.
- Theobald, D.L.I (2010), "A formal test of the theory of universal common ancestry", Nature, 465 (7295): 219–222, Bibcode:2010Natur.465..219T, doi:10.1038/nature09014, PMID 20463738
- Doolittle, W.F. (2000), "Uprooting the tree of life" (PDF), Scientific American, 282 (6): 90–95, Bibcode:2000SciAm.282b..90D, doi:10.1038/scientificamerican0200-90, PMID 10710791, archived from the original (PDF) on 31 January 2011.
- Glansdorff, N.; Xu, Y; Labedan, B. (2008), "The Last Universal Common Ancestor: Emergence, constitution and genetic legacy of an elusive forerunner", Biology Direct, 3: 29, doi:10.1186/1745-6150-3-29, PMC 2478661, PMID 18613974.
- Yoko Ohtomo; Takeshi Kakegawa; Akizumi Ishida; Toshiro Nagase; Minik T. Rosing (8 December 2013). "Evidence for biogenic graphite in early Archaean Isua metasedimentary rocks". Nature Geoscience. 7 (1): 25–28. Bibcode:2014NatGe...7...25O. doi:10.1038/ngeo2025.
- Borenstein, Seth (13 November 2013). "Oldest fossil found: Meet your microbial mom". AP News. Retrieved 15 November 2013.
- Noffke, Nora; Christian, Daniel; Wacey, David; Hazen, Robert M. (8 November 2013). "Microbially Induced Sedimentary Structures Recording an Ancient Ecosystem in the ca. 3.48 Billion-Year-Old Dresser Formation, Pilbara, Western Australia". Astrobiology. 13 (12): 1103–1124. Bibcode:2013AsBio..13.1103N. doi:10.1089/ast.2013.1030. PMC 3870916. PMID 24205812.
- Doolittle, W. Ford (2000). "Uprooting the tree of life" (PDF). Scientific American. 282 (6): 90–95. Bibcode:2000SciAm.282b..90D. doi:10.1038/scientificamerican0200-90. PMID 10710791. Archived from the original (PDF) on 7 September 2006.
- Theobald, Douglas L. (13 May 2010), "A formal test of the theory of universal common ancestry", Nature, 465 (7295): 219–222, Bibcode:2010Natur.465..219T, doi:10.1038/nature09014, ISSN 0028-0836, PMID 20463738.
- Brown, J.R.; Doolittle, W.F. (1995). "Root of the Universal Tree of Life Based on Ancient Aminoacyl-tRNA Synthetase Gene Duplications". Proc Natl Acad Sci U S A. 92 (7): 2441–2445. doi:10.1073/pnas.92.7.2441. PMC 42233. PMID 7708661.
- Gogarten, J.P.; Kibak, H.; Dittrich, P.; Taiz, L.; Bowman, E.J.; Bowman, B.J.; Manolson, M.F.; et al. (1989). "Evolution of the Vacuolar H+-ATPase: Implications for the Origin of Eukaryotes". Proc Natl Acad Sci U S A. 86 (17): 6661–6665. doi:10.1073/pnas.86.17.6661. PMC 297905. PMID 2528146.
- Gogarten, J.P.; Taiz, L. (1992). "Evolution of Proton Pumping ATPases: Rooting the Tree of Life". Photosynthesis Research. 33 (2): 137–146. doi:10.1007/BF00039176. PMID 24408574.
- Gribaldo, S; Cammarano, P (1998). "The Root of the Universal Tree of Life Inferred from Anciently Duplicated Genes Encoding Components of the Protein-Targeting Machinery". Journal of Molecular Evolution. 47 (5): 508–516. doi:10.1007/pl00006407. PMID 9797401.
- Iwabe, Naoyuki; Kuma, Kei-Ichi; Hasegawa, Masami; Osawa, Syozo; Miyata Source, Takashi; Hasegawa, Masami; Osawa, Syozo; Miyata, Takashi (1989). "Evolutionary Relationship of Archaebacteria, Eubacteria, and Eukaryotes Inferred from Phylogenetic Trees of Duplicated Genes". Proc Natl Acad Sci U S A. 86 (23): 9355–9359. doi:10.1073/pnas.86.23.9355. PMC 298494. PMID 2531898.
- Boone, David R.; Castenholz, Richard W.; Garrity, George M., eds. (2001). The Archaea and the Deeply Branching and Phototrophic Bacteria. Bergey's Manual of Systematic Bacteriology. Springer. doi:10.1007/978-0-387-21609-6. ISBN 978-0-387-21609-6.
- Valas, R.E.; Bourne, P.E. (2011). "The origin of a derived superkingdom: how a gram-positive bacterium crossed the desert to become an archaeon". Biology Direct. 6: 16. doi:10.1186/1745-6150-6-16. PMC 3056875. PMID 21356104.
- Cavalier-Smith T (2006). "Rooting the tree of life by transition analyses". Biology Direct. 1: 19. doi:10.1186/1745-6150-1-19. PMC 1586193. PMID 16834776.
- Weiss, MC; Sousa, FL; Mrnjavac, N; Neukirchen, S; Roettger, M; Nelson-Sathi, S; Martin, WF (2016). "The physiology and habitat of the last universal common ancestor". Nat Microbiol. 1 (9): 16116. doi:10.1038/NMICROBIOL.2016.116. PMID 27562259.
- Gogarten, JP; Deamer, D (November 2016). "Is LUCA a thermophilic progenitor?". Nat Microbiol. 1 (12): 16229. doi:10.1038/nmicrobiol.2016.229. PMID 27886195.
- Dacks J; Roger AJ (June 1999). "The first sexual lineage and the relevance of facultative sex". J. Mol. Evol. 48 (6): 779–783. Bibcode:1999JMolE..48..779D. doi:10.1007/PL00013156. PMID 10229582.
- Ramesh MA; Malik SB; Logsdon JM (January 2005). "A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis". Curr. Biol. 15 (2): 185–191. doi:10.1016/j.cub.2005.01.003. PMID 15668177.
- Malik SB; Pightling AW; Stefaniak LM; Schurko AM; Logsdon JM (2008). "An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis". PLOS ONE. 3 (8): e2879. Bibcode:2008PLoSO...3.2879M. doi:10.1371/journal.pone.0002879. PMC 2488364. PMID 18663385.
- Lahr DJ; Parfrey LW; Mitchell EA; Katz LA; Lara E (July 2011). "The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms". Proc. Biol. Sci. 278 (1715): 2081–2090. doi:10.1098/rspb.2011.0289. PMC 3107637. PMID 21429931.
- Chen I; Dubnau D (March 2004). "DNA uptake during bacterial transformation". Nat. Rev. Microbiol. 2 (3): 241–249. doi:10.1038/nrmicro844. PMID 15083159.
- Johnsborg O; Eldholm V; Håvarstein LS (December 2007). "Natural genetic transformation: prevalence, mechanisms and function". Res. Microbiol. 158 (10): 767–778. doi:10.1016/j.resmic.2007.09.004. PMID 17997281.
- Oklahoma State – Horizontal Gene Transfer
- Peter Gogarten. "Horizontal Gene Transfer – A New Paradigm for Biology". esalenctr.org. Retrieved 20 August 2011.
- Gibsona, Daniel G.; Benders, Gwynedd A.; Axelroda, Kevin C.; et al. (2008). "One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome". PNAS. 105 (51): 20404–20409. Bibcode:2008PNAS..10520404G. doi:10.1073/pnas.0811011106. PMC 2600582. PMID 19073939.
External links
- BBCNews: 27 September 2000, When slime is not so thick Citat: "It means that some of the lowliest creatures in the plant and animal kingdoms, such as slime and amoeba, may not be as primitive as once thought"
- SpaceRef.com, July 29, 1997: Scientists Discover Methane Ice Worms On Gulf Of Mexico Sea Floor
- The Eberly College of Science: Methane Ice Worms discovered on Gulf of Mexico Sea Floor download Publication-quality photos
- Artikel, 2000: Methane Ice Worms: Hesiocaeca methanicola. Colonizing Fossil Fuel Reserves
- SpaceRef.com, May 04, 2001: Redefining "Life as We Know it" Hesiocaeca methanicola In 1997, Charles Fisher, professor of biology at Penn State, discovered this remarkable creature living on mounds of methane ice under half a mile of ocean on the floor of the Gulf of Mexico.
- SpaceRef.com, July 29, 1997: Scientists Discover Methane Ice Worms On Gulf Of Mexico Sea Floor
- BBCNews, 18 December 2002, 'Space bugs' grown in lab Citat: "Bacillus simplex and Staphylococcus pasteuri...Engyodontium album The strains cultured by Dr Wainwright seemed to be resistant to the effects of UV – one quality required for survival in space"
- BBCNews, 19 June 2003, Ancient organism challenges cell evolution Citat: "It appears that this organelle has been conserved in evolution from prokaryotes to eukaryotes, since it is present in both"
- Interactive Syllabus for General Biology – BI 04, Saint Anselm College, Summer 2003
- Jacob Feldman: Stramenopila
- NCBI Taxonomy entry: root
- Saint Anselm College: Survey of representatives of the major Kingdoms Citat: "Number of kingdoms has not been resolved...Bacteria present a problem with their diversity...Protista present a problem with their diversity...",
- Species 2000 Indexing the world's known species. Species 2000 has the objective of enumerating all known species of plants, animals, fungi and microbes on Earth as the baseline dataset for studies of global biodiversity. It will also provide a simple access point enabling users to link from here to other data systems for all groups of organisms, using direct species-links.
- The largest organism in the world may be a fungus carpeting nearly 10 square kilometers of an Oregon forest, and may be as old as 10500 years.
- The Tree of Life
- Frequent questions from kids about life and their answers