RMND5B
| RMND5B | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | RMND5B, GID2, GID2B, required for meiotic nuclear division 5 homolog B | ||||||||||||||||||||||||
| External IDs | MGI: 1913339 HomoloGene: 100777 GeneCards: RMND5B | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | Chr 5: 178.13 – 178.15 Mb | Chr 11: 51.62 – 51.64 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Required for meiotic nuclear division 5 homolog B (S. cerevisiae), also known as RMND5B, is a protein which in humans is encoded by the RMND5B gene.[5] It has a zinc finger domain and is highly conserved throughout many eukaryotic organisms.
Protein sequence
This protein is rich in leucine (14.0%) and might belong to the protein family of leucine-rich repeats
1 meqcacvere ldkvlqkflt ygqhcersle ellhyvgqlr aelasaalqg tplsatlslv
61 msqccrkikd tvqklasdhk dihssvsrvg kaidrnfdse icgvvsdavw dareqqqqil
121 qmaivehlyq qgmlsvaeel cqestlnvdl dfkqpfleln rilealheqd lgpalewavs
181 hrqrllelns slefklhrlh firllaggpa kqlealsyar hfqpfarlhq reiqvmmgsl
241 vylrlgleks pychlldssh waeicetftr dacsllglsv esplsvsfas gcvalpvlmn
301 ikavieqrqc tgvwnhkdel pieielgmkc wyhsvfacpi lrqqtsdsnp piklicghvi
361 srdalnklin ggklkcpycp meqnpadgkr iif
Homology
CAD28476 is highly conserved in many eukaryotic organism. Its high conservation suggests that it plays a primary role in meiosis.
Orthologs
| taxonomic name | common name | NCBI entry | Percentage of sequence similarity | Length (AAs) | comments |
| Homo sapiens | Human | 100% | 393 | hypothetical Protein for Meiosis | |
| Pan troglodytes | Chimpanzee | 99.7% | 393 | required for meiotic nuclear division 5 homolog B isoform 6 | |
| Macaca mulatta | Rhesus macaque | 98.5% | 393 | Similar to CG3295-PA isoform 11 | |
| Rattus norvegicus | Norway rat | 98.2% | 393 | Similar to RIKEN cDNA 0610039K22 | |
| Mus musculus | House mouse | 97.7% | 393 | Required for meiotic nuclear division 5 homolog B | |
| Equus caballus | Horse | 97.7% | 273 | PREDICTED: similar required for meiotic nuclear division 5 homolog B | |
| Bos taurus | Cattle | 95.4% | 393 | required for meiotic nuclear division 5 homolog B | |
| Danio rerio | Zebrafish | 72.8% | 391 | required for meiotic nuclear division 5 homolog B | |
| Xenopus laevis | African clawed frog | 70% | 391 | MGC84431 protein | |
| Canis lupus familiaris | Dog | 70% | 391 | PREDICTED: similar to CG3295-PA isoform A | |
| Tetraodon nigroviridis | Spotted green pufferfish | 68.5% | 417 | unnamed protein product | |
| Ornithorhynchus anatinus | Platypus | 64.5% | 389 | ||
| Branchiostoma floridae | Florida lancelet | 57% | 491 | hypothetical protein BRAFLDRAFT_126901 | |
| Nematostella vectensis | Sea anemone | 50.1% | 389 | ||
| Culex quinquefasciatus | Southern house mosquito | 48.5% | 392 | conserved hypothetical protein | |
| Aedes aegypti | Yellow fever mosquito | 48.2% | 392 | hypothetical protein AaeL_AAEL009407 | |
| Strongylocentrotus purpuratus | Purple urchin | 48.2 | 405 | PREDICTED: Similar to MGC88921 protein | |
| Drosophila virilis | Fruit fly | 41.5% | 437 | GJ20343 | |
| Drosophila melanogaster | Fruit fly | 40.3% | 431 | CG3295 | |
| Acyrthosiphon pisum | Acrythosiphon | 39.5% | 366 | PREDICTED: required for meiothic nuclear division 5 homolg A | |
| Oryza sativa | Rice | 32.2% | 386 | membrane protein-like | |
Zinc finger domain
Two domains were predicted by the program BLIMPS[6] to exist in the protein of which one of the domains contains a zinc finger domain.
Zinc finger domains assist the binding of the protein to nucleic acids. This points to a direct interaction of CAD28476 with DNA during meiosis. By comparing CAD28476 with a related zinc finger protein[7] in a local sequence alignment using LALIGN,[8] the amino acids His359, Cys381 and Cys384 could be attributed to the zinc finger domain. This zinc finger structure is uncommon in the way that it involves one histidine instead of two.
Expression
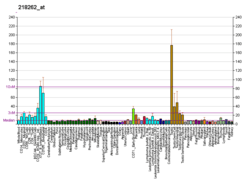
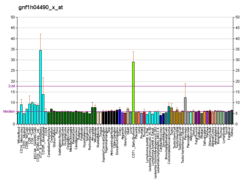
Microarray data show that CAD28476 is highly expressed in tissue where meiosis occurs like in testis and ovaries. Moreover it is also highly expressed in the brain around the hypothalamus.
Transcriptional regulation
The analysis of the promoter region (tools on the page rVista .[9] were used) shows that there are several transcription factor binding sites localized in conserved regions . It is very likely that the transcription factor Ets-1 which belongs to the ETS transcription factor family and its core binding factor CBF are involved in regulation of transcription since they both have independent binding sites.
Interacting proteins
There were two proteins predicted[10] which interact with CAD28476.
| name of the protein | Uniprot entry | information about the gene |
|---|---|---|
| Ewing sarcoma breakpoint region 1, isoform CRA_a | gene encodes a putative RNA binding protein. Mutations in this gene, specifically a t(11;22)(q24;q12) translocation, are known to cause Ewing sarcoma as well as neuroectodermal and various other tumors. | |
| Mothers against decapentaplegic homolog 4 | SMAD4 |
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000145916 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000001054 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ "Entrez Gene: RMND5B required for meiotic nuclear division 5 homolog B (S. cerevisiae)".
- ↑ Jorja Henikoff, Fred Hutchinson Cancer Research Center, 1100 Fairview AV N, A1-162, PO Box 19024 Seattle, WA 98109-1024 FAX: 206-667-5889
- ↑ "PDB entry of 2ct2". Retrieved 12 May 2009.
- ↑ © 1997 by William R. Pearson and the University of Virginia (This is from distribution "fasta20u66", version 2.0u66, Sep., 1998, sale or incorporation into a commercial product expressly forbidden without permission)
- ↑ "rvista Home Page". Retrieved 12 May 2009.
- ↑ "homepage of Genecard". Retrieved 12 May 2009.
Further reading
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Clark HF, Gurney AL, Abaya E, et al. (2003). "The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment". Genome Res. 13 (10): 2265–70. doi:10.1101/gr.1293003. PMC 403697. PMID 12975309.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Colland F, Jacq X, Trouplin V, et al. (2004). "Functional proteomics mapping of a human signaling pathway". Genome Res. 14 (7): 1324–32. doi:10.1101/gr.2334104. PMC 442148. PMID 15231748.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Pope SN, Lee IR (2005). "Yeast two-hybrid identification of prostatic proteins interacting with human sex hormone-binding globulin". J. Steroid Biochem. Mol. Biol. 94 (1–3): 203–8. doi:10.1016/j.jsbmb.2005.01.007. PMID 15862967.
- Rual JF, Venkatesan K, Hao T, et al. (2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.