Protein structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a residue indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein.[1] To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, and dual polarisation interferometry to determine the structure of proteins.
Protein structures range in size from tens to several thousand amino acids.[2] By physical size, proteins are classified as nanoparticles, between 1–100 nm. Very large aggregates can be formed from protein subunits. For example, many thousands of actin molecules assemble into a microfilament.
A protein generally undergo reversible structural changes in performing its biological function. The alternative structures of the same protein are referred to as different conformational isomers, or simply, conformations, and transitions between them are called conformational changes.
Levels of protein structure
There are four distinct levels of protein structure.
Primary structure
The primary structure of a protein refers to the sequence of amino acids in the polypeptide chain. The primary structure is held together by peptide bonds that are made during the process of protein biosynthesis. The two ends of the polypeptide chain are referred to as the carboxyl terminus (C-terminus) and the amino terminus (N-terminus) based on the nature of the free group on each extremity. Counting of residues always starts at the N-terminal end (NH2-group), which is the end where the amino group is not involved in a peptide bond. The primary structure of a protein is determined by the gene corresponding to the protein. A specific sequence of nucleotides in DNA is transcribed into mRNA, which is read by the ribosome in a process called translation. The sequence of amino acids in insulin was discovered by Frederick Sanger, establishing that proteins have defining amino acid sequences.[3][4] The sequence of a protein is unique to that protein, and defines the structure and function of the protein. The sequence of a protein can be determined by methods such as Edman degradation or tandem mass spectrometry. Often, however, it is read directly from the sequence of the gene using the genetic code. It is strictly recommended to use the words "amino acid residues" when discussing proteins because when a peptide bond is formed, a water molecule is lost, and therefore proteins are made up of amino acid residues. Post-translational modification such as disulfide bond formation, phosphorylations and glycosylations are usually also considered a part of the primary structure, and cannot be read from the gene. For example, insulin is composed of 51 amino acids in 2 chains. One chain has 31 amino acids, and the other has 20 amino acids.
Secondary structure
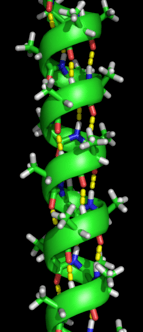
Secondary structure refers to highly regular local sub-structures on the actual polypeptide backbone chain. Two main types of secondary structure, the α-helix and the β-strand or β-sheets, were suggested in 1951 by Linus Pauling and coworkers.[5] These secondary structures are defined by patterns of hydrogen bonds between the main-chain peptide groups. They have a regular geometry, being constrained to specific values of the dihedral angles ψ and φ on the Ramachandran plot. Both the α-helix and the β-sheet represent a way of saturating all the hydrogen bond donors and acceptors in the peptide backbone. Some parts of the protein are ordered but do not form any regular structures. They should not be confused with random coil, an unfolded polypeptide chain lacking any fixed three-dimensional structure. Several sequential secondary structures may form a "supersecondary unit".[6]
Tertiary structure
Tertiary structure refers to the three-dimensional structure of monomeric and multimeric protein molecules. The α-helixes and β-pleated-sheets are folded into a compact globular structure. The folding is driven by the non-specific hydrophobic interactions, the burial of hydrophobic residues from water, but the structure is stable only when the parts of a protein domain are locked into place by specific tertiary interactions, such as salt bridges, hydrogen bonds, and the tight packing of side chains and disulfide bonds. The disulfide bonds are extremely rare in cytosolic proteins, since the cytosol (intracellular fluid) is generally a reducing environment.
Quaternary structure
Quaternary structure is the three-dimensional structure consisting of the aggregation of two or more individual polypeptide chains (subunits) that operate as a single functional unit (multimer). The resulting multimer is stabilized by the same non-covalent interactions and disulfide bonds as in tertiary structure. There are many possible quaternary structure organisations.[7] Complexes of two or more polypeptides (i.e. multiple subunits) are called multimers. Specifically it would be called a dimer if it contains two subunits, a trimer if it contains three subunits, a tetramer if it contains four subunits, and a pentamer if it contains five subunits. The subunits are frequently related to one another by symmetry operations, such as a 2-fold axis in a dimer. Multimers made up of identical subunits are referred to with a prefix of "homo-" (e.g. a homotetramer) and those made up of different subunits are referred to with a prefix of "hetero-", for example, a heterotetramer, such as the two alpha and two beta chains of hemoglobin.
Domains, motifs, and folds in protein structure
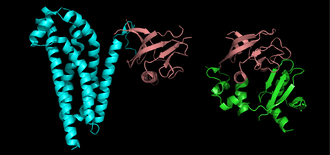
Proteins are frequently described as consisting of several structural units. These units include domains, motifs, and folds. Despite the fact that there are about 100,000 different proteins expressed in eukaryotic systems, there are many fewer different domains, structural motifs and folds.
Structural domain
A structural domain is an element of the protein's overall structure that is self-stabilizing and often folds independently of the rest of the protein chain. Many domains are not unique to the protein products of one gene or one gene family but instead appear in a variety of proteins. Domains often are named and singled out because they figure prominently in the biological function of the protein they belong to; for example, the "calcium-binding domain of calmodulin". Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimera proteins.
Structural and sequence motif
The structural and sequence motifs refer to short segments of protein three-dimensional structure or amino acid sequence that were found in a large number of different proteins.
Supersecondary structure
The supersecondary structure refers to a specific combination of secondary structure elements, such as β-α-β units or a helix-turn-helix motif. Some of them may be also referred to as structural motifs.
Protein fold
A protein fold refers to the general protein architecture, like a helix bundle, β-barrel, Rossman fold or different "folds" provided in the Structural Classification of Proteins database.[8] A related concept is protein topology that refers to the arrangement of contacts within the protein.
Superdomain
A superdomain consists of two or more nominally unrelated structural domains that are inherited as a single unit and occur in different proteins.[9] An example is provided by the protein tyrosine phosphatase domain and C2 domain pair in PTEN, several tensin proteins, auxilin and proteins in plants and fungi. The PTP-C2 superdomain evidently came into existence prior to the divergence of fungi, plants and animals is therefore likely to be about 1.5 billion years old.
Protein folding
As it is translated, polypeptides exit the ribosome as a random coil and folds into its native state.[10][11] Since the fold is determined by a network of interactions between amino acids in the polypeptide, the final structure of the protein chain is determined by its amino acid sequence (Anfinsen's dogma).[12]
Protein stability
Protein stability depends upon a few factors such as 1) Non-covalent electrostatic interactions 2) Hydrophobic interactions These interaction energies are from the order of 20-40 kJ/mol. Proteins are very sensitive to changing temperatures and a change in temperature may result in unfolding or denaturation. Protein denaturation may result in loss of function, and loss of native state.
X-ray crystallography and calorimetry indicates that there is no general mechanism that describes the effect of temperature change on the functions and structure of proteins. This is due to the fact that proteins do not represent a uniform class of chemical entities from an energetic point of view. The structure and stability of an individual protein depends on the ratio of its polar and non-polar residues. They contribute to the conformational and the net enthalpies of local and non-local interactions.
Taking the weak intermolecular interactions responsible for structural integrity into consideration, it is hard to predict the effects of temperature because there are too many unknown factors contributing to the hypothetical free energy balance and its temperature dependence. Internal salt linkages produce thermal stability, and whether cold temperature results in the destabilization of these linkages is unknown.
In principle, the free energy of stabilization of soluble globular proteins does not exceed 50-100 kJ/mol. The stabilization is based on the equivalent of few hydrogen bonds, ion pairs, or hydrophobic interactions, even though numerous intramolecular interactions results in stabilization. Taking into consideration the large number of hydrogen bonds that take place for the stabilization of secondary structures, and the stabilization of the inner core through hydrophobic interactions, the free energy of stabilization emerges as small difference between large numbers. Therefore, the structure of a native protein is not optimized for the maximum stability.[13]
Protein structure determination
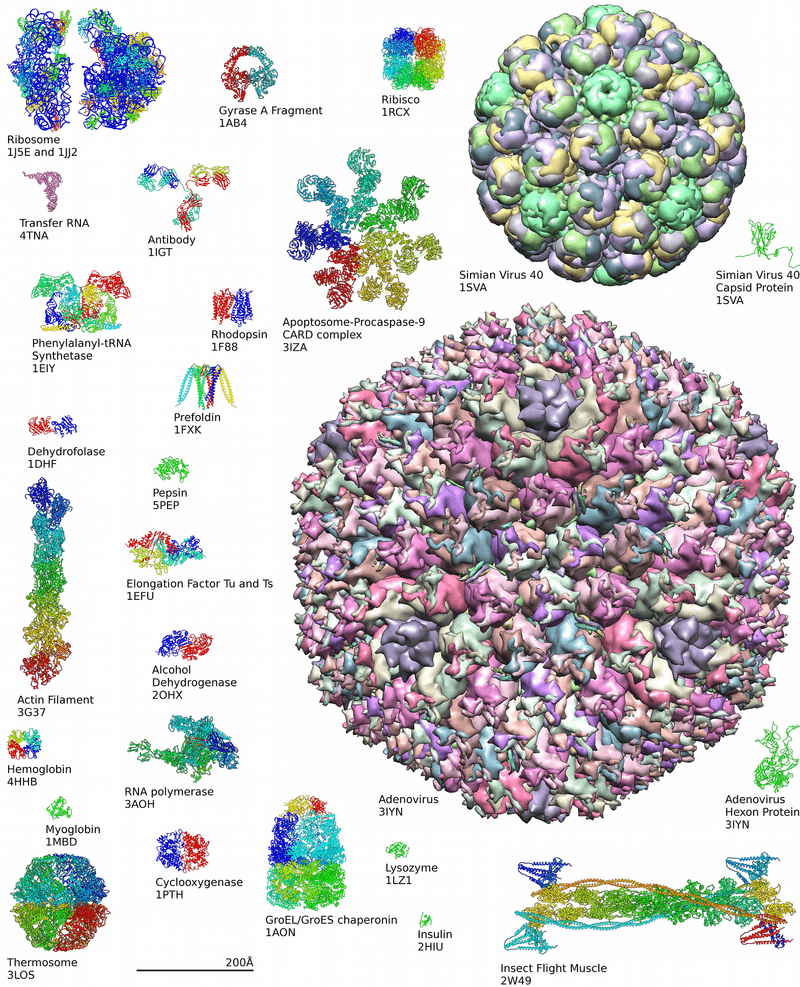

Around 90% of the protein structures available in the Protein Data Bank have been determined by X-ray crystallography.[14] This method allows one to measure the three-dimensional (3-D) density distribution of electrons in the protein, in the crystallized state, and thereby infer the 3-D coordinates of all the atoms to be determined to a certain resolution. Roughly 9% of the known protein structures have been obtained by nuclear magnetic resonance (NMR) techniques. For larger protein complexes, cryo-electron microscopy can determine protein structures. The resolution is typically lower than that of X-ray crystallography, or NMR, but the maximum resolution is steadily increasing. This technique is still a particularly valuable for very large protein complexes such as virus coat proteins and amyloid fibers.
General secondary structure composition can be determined via circular dichroism. Vibrational spectroscopy can also be used to characterize the conformation of peptides, polypeptides, and proteins.[15] Two-dimensional infrared spectroscopy has become a valuable method to investigate the structures of flexible peptides and proteins that cannot be studied with other methods.[16][17] A more qualitative picture of protein structure is often obtained by proteolysis, which is also useful to screen for more crystallizable protein samples. Novel implementations of this approach, including fast parallel proteolysis (FASTpp), can probe the structured fraction and its stability without the need for purification.[18] Once a protein's structure has been experimentally determined, further detailed studies can be done computationally, using molecular dynamic simulations of that structure.[19]
Protein Sequence Analysis: Ensembles

Proteins are often thought of as relatively stable structures that have a set tertiary structure and experience conformational changes as a result of being modified by other proteins or as part of enzymatic activity. However proteins have varying degrees of stability and some of the less stable variants are intrinsically disordered proteins. These proteins exist and function in a relatively 'disordered' state lacking a stable tertiary structure. As a result, they are difficult to describe in a standard protein structure model that was designed for proteins with a fixed tertiary structure. Conformational ensembles have been devised as a way to provide a more accurate and 'dynamic' representation of the conformational state of intrinsically disordered proteins. Conformational ensembles function by attempting to represent the various conformations of intrinsically disordered proteins within an ensemble file (the type found at the Protein Ensemble Database).
Protein ensemble files are a representation of a protein that can be considered to have a flexible structure. Creating these files requires determining which of the various theoretically possible protein conformations actually exist. One approach is to apply computational algorithms to the protein data in order to try to determine the most likely set of conformations for an ensemble file.
There are multiple methods for preparing data for the Protein Ensemble Database that fall into two general methodologies – pool and molecular dynamics (MD) approaches (diagrammed in the figure). The pool based approach uses the protein’s amino acid sequence to create a massive pool of random conformations. This pool is then subjected to more computational processing that creates a set of theoretical parameters for each conformation based on the structure. Conformational subsets from this pool whose average theoretical parameters closely match known experimental data for this protein are selected.[20]
The molecular dynamics approach takes multiple random conformations at a time and subjects all of them to experimental data. Here the experimental data is serving as limitations to be placed on the conformations (e.g. known distances between atoms). Only conformations that manage to remain within the limits set by the experimental data are accepted. This approach often applies large amounts of experimental data to the conformations which is a very computationally demanding task.[20]
| Protein | Data Type | Protocol | PED ID | References |
|---|---|---|---|---|
| Sic1/Cdc4 | NMR and SAXS | Pool-based | PED9AAA | [21] |
| p15 PAF | NMR and SAXS | Pool-based | PED6AAA | [22] |
| MKK7 | NMR | Pool-based | PED5AAB | [23] |
| Beta-synuclein | NMR | MD-based | PED1AAD | [24] |
| P27 KID | NMR | MD-based | PED2AAA | [25] |
(adapted from image in "Computational approaches for inferring the functions of intrinsically disordered proteins"[20])
Protein structure databases
Protein structure database is a database that is modeled around the various experimentally determined protein structures. The aim of most protein structure databases is to organize and annotate the protein structures, providing the biological community access to the experimental data in a useful way. Data included in protein structure databases often includes 3D coordinates as well as experimental information, such as unit cell dimensions and angles for x-ray crystallography determined structures. Though most instances, in this case either proteins or a specific structure determinations of a protein, also contain sequence information and some databases even provide means for performing sequence based queries, the primary attribute of a structure database is structural information, whereas sequence databases focus on sequence information, and contain no structural information for the majority of entries. Protein structure databases are critical for many efforts in computational biology such as structure based drug design, both in developing the computational methods used and in providing a large experimental dataset used by some methods to provide insights about the function of a protein.[26]
Structure classification
Protein structures can be grouped based on their similarity or a common evolutionary origin. The Structural Classification of Proteins database[27] and CATH database[28] provide two different structural classifications of proteins. Shared structure between proteins is considered evidence of evolutionary relatedness between proteins and is used group proteins together into protein superfamilies.[29]
Computational prediction of protein structure
The generation of a protein sequence is much easier than the determination of a protein structure. However, the structure of a protein gives much more insight in the function of the protein than its sequence. Therefore, a number of methods for the computational prediction of protein structure from its sequence have been developed.[30] Ab initio prediction methods use just the sequence of the protein. Threading and homology modeling methods can build a 3-D model for a protein of unknown structure from experimental structures of evolutionarily-related proteins, called a protein family.
See also
- 3did
- Biomolecular structure
- Gene structure
- Nucleic acid structure
- Z-matrix conversion from Torsion angles to Cartesian coordinates
References
- ↑ H. Stephen Stoker (1 January 2015). Organic and Biological Chemistry. Cengage Learning. p. 371. ISBN 978-1-305-68645-8.
- ↑ Brocchieri L, Karlin S (2005-06-10). "Protein length in eukaryotic and prokaryotic proteomes". Nucleic Acids Research. 33 (10): 3390–3400. doi:10.1093/nar/gki615. PMC 1150220. PMID 15951512.
- ↑ Sanger, F.; Tuppy, H. (1951-09-01). "The amino-acid sequence in the phenylalanyl chain of insulin. I. The identification of lower peptides from partial hydrolysates". The Biochemical Journal. 49 (4): 463–481. doi:10.1042/bj0490463. ISSN 0264-6021. PMC 1197535. PMID 14886310.
- ↑ Sanger, F. (1959-05-15). "Chemistry of Insulin". Science. 129 (3359): 1340–1344. doi:10.1126/science.129.3359.1340. ISSN 0036-8075. PMID 13658959.
- ↑ Pauling L, Corey RB, Branson HR (1951). "The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain". Proc Natl Acad Sci USA. 37 (4): 205–211. doi:10.1073/pnas.37.4.205. PMC 1063337. PMID 14816373.
- ↑ Chiang YS, Gelfand TI, Kister AE, Gelfand IM (2007). "New classification of supersecondary structures of sandwich-like proteins uncovers strict patterns of strand assemblage". Proteins. 68 (4): 915–921. doi:10.1002/prot.21473. PMID 17557333.
- ↑ Moutevelis, Efrosini; Woolfson, Derek N. (2009-01). "A Periodic Table of Coiled-Coil Protein Structures". Journal of Molecular Biology. 385 (3): 726–732. doi:10.1016/j.jmb.2008.11.028. ISSN 0022-2836. Check date values in:
|date=(help) - ↑ Govindarajan S, Recabarren R, Goldstein RA (17 September 1999). "Estimating the total number of protein folds". Proteins. 35 (4): 408–414. doi:10.1002/(SICI)1097-0134(19990601)35:4<408::AID-PROT4>3.0.CO;2-A. PMID 10382668.
- ↑ Haynie DT, Xue B (2015). "Superdomain in the protein structure hierarchy: the case of PTP-C2". Protein Science. 24: 874–82. doi:10.1002/pro.2664. PMC 4420535. PMID 25694109.
- ↑ "Folding at the birth of the nascent chain: coordinating translation with co-translational folding". Current Opinion in Structural Biology. 21 (1): 25–31. 2011-02-01. doi:10.1016/j.sbi.2010.10.008. ISSN 0959-440X.
- ↑ Alberts, Bruce; Alexander Johnson; Julian Lewis; Martin Raff; Keith Roberts; Peter Walters (2002). "The Shape and Structure of Proteins". Molecular Biology of the Cell; Fourth Edition. New York and London: Garland Science. ISBN 0-8153-3218-1.
- ↑ Anfinsen, C. (1972). "The formation and stabilization of protein structure". Biochem. J. 128 (4): 737–49. doi:10.1042/bj1280737. PMC 1173893. PMID 4565129.
- ↑ Jaenicke, R.; Heber, U.; Franks, F.; Chapman, D.; Griffin, Mary C. A.; Hvidt, A.; Cowan, D. A. (1990). "Protein Structure and Function at Low Temperatures [and Discussion]". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 326 (1237): 535–553. JSTOR 2398703.
- ↑ Kendrew, J.C.; Bodo, G.; Dintzis, H. M.; Parrish, R. G.; Wyckoff, H.; Phillips, D.C. (1958). "A Three-Dimensional Model of the Myoglobin Molecule Obtained by X-Ray Analysis". Nature. 181: 662–666. doi:10.1038/181662a0. PMID 13517261.
- ↑ Krimm, Samuel; Bandekar, J. (1986). "Vibrational Spectroscopy and Conformation of Peptides, Polypeptides, and Proteins". Advances in Protein Chemistry. Advances in Protein Chemistry. 38 (C): 181–364. doi:10.1016/S0065-3233(08)60528-8. ISBN 9780120342389.
- ↑ Lessing, J.; Roy, S.; Reppert, M.; Baer, M.; Marx, D.; Jansen, T.L.C.; Knoester, J.; Tokmakoff, A. (2012). "Identifying Residual Structure in Intrinsically Disordered Systems: A 2D IR Spectroscopic Study of the GVGXPGVG Peptide". J. Am. Chem. Soc. 134: 5032–5035. doi:10.1021/ja2114135. PMID 22356513.
- ↑ Jansen, T.L.C.; Knoester, J. (2008). "Two-dimensional infrared population transfer spectroscopy for enhancing structural markers of proteins". Biophys. J. 94: 1818–1825. doi:10.1529/biophysj.107.118851. PMC 2242754. PMID 17981904.
- ↑ Minde DP, Maurice MM, Rüdiger SG (2012). Uversky, Vladimir N, ed. "Determining biophysical protein stability in lysates by a fast proteolysis assay, FASTpp". PLoS ONE. 7 (10): e46147. doi:10.1371/journal.pone.0046147. PMC 3463568. PMID 23056252.
- ↑ Kumari I, Sandhu P, Ahmed M, Akhter Y. Molecular Dynamics Simulations, Challenges and Opportunities: A Biologist's Prospective. Curr Protein Pept Sci. 2017 Aug 30;18(11):1163-1179. doi: 10.2174/1389203718666170622074741. Link for preprint.
- 1 2 3 4 Varadi, Mihaly; Vranken, Wim; Guharoy, Mainak; Tompa, Peter (2015-01-01). "Computational approaches for inferring the functions of intrinsically disordered proteins". Frontiers in Molecular Biosciences. 2: 45. doi:10.3389/fmolb.2015.00045. PMC 4525029. PMID 26301226.
- ↑ Mittag, Tanja; Marsh, Joseph; Grishaev, Alexander; Orlicky, Stephen; Lin, Hong; Sicheri, Frank; Tyers, Mike; Forman-Kay, Julie D. (2010-03-14). "Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase". Structure. 18 (4): 494–506. doi:10.1016/j.str.2010.01.020. ISSN 1878-4186. PMC 2924144. PMID 20399186.
- ↑ De Biasio, Alfredo; Ibáñez de Opakua, Alain; Cordeiro, Tiago N.; Villate, Maider; Merino, Nekane; Sibille, Nathalie; Lelli, Moreno; Diercks, Tammo; Bernadó, Pau (2014-02-18). "p15PAF is an intrinsically disordered protein with nonrandom structural preferences at sites of interaction with other proteins". Biophysical Journal. 106 (4): 865–874. doi:10.1016/j.bpj.2013.12.046. ISSN 1542-0086. PMC 3944474. PMID 24559989.
- ↑ Kragelj, Jaka; Palencia, Andrés; Nanao, Max H.; Maurin, Damien; Bouvignies, Guillaume; Blackledge, Martin; Jensen, Malene Ringkjøbing (2015-03-17). "Structure and dynamics of the MKK7-JNK signaling complex". Proceedings of the National Academy of Sciences of the United States of America. 112 (11): 3409–3414. doi:10.1073/pnas.1419528112. ISSN 1091-6490. PMC 4371970. PMID 25737554.
- ↑ Allison, Jane R.; Rivers, Robert C.; Christodoulou, John C.; Vendruscolo, Michele; Dobson, Christopher M. (2014-11-25). "A relationship between the transient structure in the monomeric state and the aggregation propensities of α-synuclein and β-synuclein". Biochemistry. 53 (46): 7170–7183. doi:10.1021/bi5009326. ISSN 1520-4995. PMC 4245978. PMID 25389903.
- ↑ Sivakolundu, Sivashankar G.; Bashford, Donald; Kriwacki, Richard W. (2005-11-11). "Disordered p27Kip1 exhibits intrinsic structure resembling the Cdk2/cyclin A-bound conformation". Journal of Molecular Biology. 353 (5): 1118–1128. doi:10.1016/j.jmb.2005.08.074. ISSN 0022-2836. PMID 16214166.
- ↑ Laskowski, RA. "Protein structure databases". Mol Biotechnol. PMID 21225378.
- ↑ Murzin, A. G.; Brenner, S.; Hubbard, T.; Chothia, C. (1995). "SCOP: A structural classification of proteins database for the investigation of sequences and structures" (PDF). Journal of Molecular Biology. 247 (4): 536–540. doi:10.1016/S0022-2836(05)80134-2. PMID 7723011. Archived from the original (PDF) on 26 April 2012.
- ↑ Orengo, C. A.; Michie, A. D.; Jones, S.; Jones, D. T.; Swindells, M. B.; Thornton, J. M. (1997). "CATH--a hierarchic classification of protein domain structures". Structure. 5 (8): 1093–1108. doi:10.1016/S0969-2126(97)00260-8. PMID 9309224.
- ↑ Holm, L; Rosenström, P (July 2010). "Dali server: conservation mapping in 3D". Nucleic Acids Research. 38 (Web Server issue): W545–9. doi:10.1093/nar/gkq366. PMC 2896194. PMID 20457744.
- ↑ Zhang Y (2008). "Progress and challenges in protein structure prediction". Curr Opin Struct Biol. 18 (3): 342–348. doi:10.1016/j.sbi.2008.02.004. PMC 2680823. PMID 18436442.
.png)