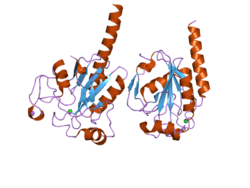GPX3
Glutathione peroxidase 3 (GPx-3), also known as plasma glutathione peroxidase (GPx-P) or extracellular glutathione peroxidase is an enzyme that in humans is encoded by the GPX3 gene.[5][6][7]
GPx-3 belongs to the glutathione peroxidase family, which functions in the detoxification of hydrogen peroxide. It contains a selenocysteine (Sec) residue at its active site. The selenocysteine is encoded by the UGA codon, which normally signals translation termination. The 3' UTR of Sec-containing genes have a common stem-loop structure, the sec insertion sequence (SECIS), which is necessary for the recognition of UGA as a Sec codon rather than as a stop signal.[5]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000211445 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000018339 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- 1 2 "Entrez Gene: glutathione peroxidase 3 (plasma)".
- ↑ Takahashi K, Avissar N, Whitin J, Cohen H (August 1987). "Purification and characterization of human plasma glutathione peroxidase: a selenoglycoprotein distinct from the known cellular enzyme". Arch. Biochem. Biophys. 256 (2): 677–86. doi:10.1016/0003-9861(87)90624-2. PMID 3619451.
- ↑ Chu FF (1994). "The human glutathione peroxidase genes GPX2, GPX3, and GPX4 map to chromosomes 14, 5, and 19, respectively". Cytogenet. Cell Genet. 66 (2): 96–8. doi:10.1159/000133675. PMID 8287691.
Further reading
- Guey LT, García-Closas M, Murta-Nascimento C, et al. (2010). "Genetic susceptibility to distinct bladder cancer subphenotypes". Eur. Urol. 57 (2): 283–92. doi:10.1016/j.eururo.2009.08.001. PMC 3220186. PMID 19692168.
- Lin JC, Kuo WR, Chiang FY, et al. (2009). "Glutathione peroxidase 3 gene polymorphisms and risk of differentiated thyroid cancer". Surgery. 145 (5): 508–13. doi:10.1016/j.surg.2008.12.008. PMID 19375609.
- Holtzman JL (2002). "The role of low levels of the serum glutathione-dependent peroxidase and glutathione and high levels of serum homocysteine in the development of cardiovascular disease". Clin. Lab. 48 (3–4): 129–30. PMID 11934214.
- Fullerton JM, Tiwari Y, Agahi G, et al. (2010). "Assessing oxidative pathway genes as risk factors for bipolar disorder". Bipolar Disord. 12 (5): 550–6. doi:10.1111/j.1399-5618.2010.00834.x. PMID 20712757.
- Hosgood HD, Zhang L, Shen M, et al. (2009). "Association between genetic variants in VEGF, ERCC3 and occupational benzene haematotoxicity". Occup Environ Med. 66 (12): 848–53. doi:10.1136/oem.2008.044024. PMC 2928224. PMID 19773279.
- Liang XS, Caporaso N, McMaster ML, et al. (2009). "Common genetic variants in candidate genes and risk of familial lymphoid malignancies". Br. J. Haematol. 146 (4): 418–23. doi:10.1111/j.1365-2141.2009.07790.x. PMC 2890251. PMID 19573080.
- Hosgood HD, Menashe I, He X, et al. (2009). "PTEN identified as important risk factor of chronic obstructive pulmonary disease". Respir Med. 103 (12): 1866–70. doi:10.1016/j.rmed.2009.06.016. PMC 2783799. PMID 19625176.
- Mistry HD, Kurlak LO, Williams PJ, et al. (2010). "Differential expression and distribution of placental glutathione peroxidases 1, 3 and 4 in normal and preeclamptic pregnancy". Placenta. 31 (5): 401–8. doi:10.1016/j.placenta.2010.02.011. PMID 20303587.
- Hosgood HD, Menashe I, Shen M, et al. (2008). "Pathway-based evaluation of 380 candidate genes and lung cancer susceptibility suggests the importance of the cell cycle pathway". Carcinogenesis. 29 (10): 1938–43. doi:10.1093/carcin/bgn178. PMC 2722857. PMID 18676680.
- Iida R, Tsubota E, Yuasa I, et al. (2009). "Simultaneous genotyping of 11 non-synonymous SNPs in the 4 glutathione peroxidase genes using the multiplex single base extension method". Clin. Chim. Acta. 402 (1–2): 79–82. doi:10.1016/j.cca.2008.12.027. PMID 19161995.
- Talmud PJ, Drenos F, Shah S, et al. (2009). "Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip". Am. J. Hum. Genet. 85 (5): 628–42. doi:10.1016/j.ajhg.2009.10.014. PMC 2775832. PMID 19913121.
- Wang JY, Yang IP, Wu DC, et al. (2010). "Functional glutathione peroxidase 3 polymorphisms associated with increased risk of Taiwanese patients with gastric cancer". Clin. Chim. Acta. 411 (19–20): 1432–6. doi:10.1016/j.cca.2010.05.026. PMID 20576521.
- Westphal K, Stangl V, Fähling M, et al. (2009). "Human-specific induction of glutathione peroxidase-3 by proteasome inhibition in cardiovascular cells". Free Radic. Biol. Med. 47 (11): 1652–60. doi:10.1016/j.freeradbiomed.2009.09.017. PMID 19766714.
- Shen M, Vermeulen R, Rajaraman P, et al. (2009). "Polymorphisms in innate immunity genes and lung cancer risk in Xuanwei, China". Environ. Mol. Mutagen. 50 (4): 285–90. doi:10.1002/em.20452. PMC 2666781. PMID 19170196.
- Saga Y, Ohwada M, Suzuki M, et al. (2008). "Glutathione peroxidase 3 is a candidate mechanism of anticancer drug resistance of ovarian clear cell adenocarcinoma". Oncol. Rep. 20 (6): 1299–303. doi:10.3892/or_00000144. PMID 19020706.
- Zhang X, Yang JJ, Kim YS, et al. (2010). "An 8-gene signature, including methylated and down-regulated glutathione peroxidase 3, of gastric cancer". Int. J. Oncol. 36 (2): 405–14. doi:10.3892/ijo_00000513. PMID 20043075.
- Chung SS, Kim M, Youn BS, et al. (2009). "Glutathione peroxidase 3 mediates the antioxidant effect of peroxisome proliferator-activated receptor gamma in human skeletal muscle cells". Mol. Cell. Biol. 29 (1): 20–30. doi:10.1128/MCB.00544-08. PMC 2612482. PMID 18936159.
- Wang Y, Fu W, Xie F, et al. (2010). "Common polymorphisms in ITGA2, PON1 and THBS2 are associated with coronary atherosclerosis in a candidate gene association study of the Chinese Han population". J. Hum. Genet. 55 (8): 490–4. doi:10.1038/jhg.2010.53. PMID 20485444.
- Moyer AM, Sun Z, Batzler AJ, et al. (2010). "Glutathione pathway genetic polymorphisms and lung cancer survival after platinum-based chemotherapy". Cancer Epidemiol. Biomarkers Prev. 19 (3): 811–21. doi:10.1158/1055-9965.EPI-09-0871. PMC 2837367. PMID 20200426.
- Bailey SD, Xie C, Do R, et al. (2010). "Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study". Diabetes Care. 33 (10): 2250–3. doi:10.2337/dc10-0452. PMC 2945168. PMID 20628086.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.