G-quadruplex
In molecular biology, G-quadruplex secondary structures[1] are formed in nucleic acids by sequences that are rich in guanine. They are helical structures containing guanine tetrads that can form from one,[2] two[3] or four strands.[4] The unimolecular forms often occur naturally near the ends of the chromosomes, better known as the telomeric regions, and in transcriptional regulatory regions of multiple genes and oncogenes.[5] Four guanine bases can associate through Hoogsteen hydrogen bonding to form a square planar structure called a guanine tetrad (also called G-tetrad or G-quartet), and two or more guanine tetrads can stack on top of each other to form a G-quadruplex. The placement and bonding to form G-quadruplexes are not random and serve very unusual functional purposes. The quadruplex structure is further stabilized by the presence of a cation, especially potassium, which sits in a central channel between each pair of tetrads.[2] They can be formed of DNA, RNA, LNA, and PNA, and may be intramolecular, bimolecular, or tetramolecular.[6] Depending on the direction of the strands or parts of a strand that form the tetrads, structures may be described as parallel or antiparallel. G-quadruplex structures can be computationally predicted from DNA or RNA sequence motifs, but their actual structures can be quite varied within and between the motifs, which can number over 100,000 per genome. Their activities in basic genetic processes are an active area of research in telomere, gene regulation, and functional genomics research.[7]
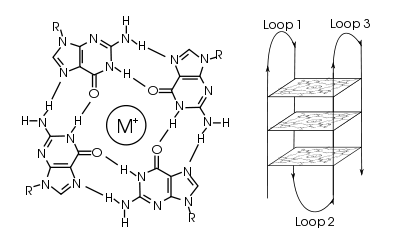

Quadruplex topology
The length of the nucleic acid sequences involved in tetrad formation determines how the quadruplex folds. Short sequences, consisting of only a single contiguous run of three or more guanine bases, require four individual strands to form a quadruplex. Such a quadruplex is described as tetramolecular, reflecting the requirement of four separate strands. The term G4 DNA was originally reserved for these tetramolecular structures that might play a role in meiosis.[4] However, as currently used in molecular biology, the term G4 can mean G-quadruplexes of any molecularity. Longer sequences, which contain two contiguous runs of three or more guanine bases, where the guanine regions are separated by one or more bases, only require two such sequences to provide enough guanine bases to form a quadruplex. These structures, formed from two separate G-rich strands, are termed bimolecular quadruplexes. Finally, sequences which contain four distinct runs of guanine bases can form stable quadruplex structures by themselves, and a quadruplex formed entirely from a single strand is called an intramolecular quadruplex.[8]
Depending on how the individual runs of guanine bases are arranged in a bimolecular or intramolecular quadruplex, a quadruplex can adopt one of a number of topologies with varying loop configurations.[9] If all strands of DNA proceed in the same direction, the quadruplex is termed parallel. For intramolecular quadruplexes, this means that any loop regions present must be of the propeller type, positioned to the sides of the quadruplex. If one or more of the runs of guanine bases has a 5’-3’ direction opposite to the other runs of guanine bases, the quadruplex is said to have adopted an antiparallel topology. The loops joining runs of guanine bases in intramolecular antiparallel quadruplexes are either diagonal, joining two diagonally opposite runs of guanine bases, or lateral (edgewise) type loops, joining two adjacent runs of guanine base pairs.
In quadruplexes formed from double-stranded DNA, possible interstrand topologies have also been discussed [10] .[11] Interstrand quadruplexes contain guanines that originate from both strands of dsDNA.
Structure and functional role in genome
Following sequencing of the human genome, many guanine-rich sequences that had the potential to form quadraplexes were discovered.[12] Depending on cell type and cell cycle, mediating factors such as DNA-binding proteins like chromatin, composed of DNA tightly wound around histone proteins, and other environmental conditions and stresses affect the dynamic formation of quadraplexes. For instance, quantitative assessments of the thermodynamics of molecular crowding indicate that the antiparallel g-quadruplex is stabilized by molecular crowding.[13] This effect seems to be mediated by alteration of the hydration of the DNA and its effect on Hoogsteen base pair bonding.[14] These quadruplexes seemed to readily occur at the ends of chromosome. In addition, the propensity of g-quadruplex formation during transcription in RNA sequences with the potential to form mutually exclusive hairpin or G-quadruplex structures depends heavily on the position of the hairpin-forming sequence.[15]
Because repair enzymes would naturally recognize ends of linear chromosomes as damaged DNA and would process them as such to harmful effect for the cell, clear signaling and tight regulation is needed at the ends of linear chromosomes. Telomeres function to provide this signaling. Telomeres, rich in guanine and with a propensity to form g-quadruplexes, are located at the terminal ends of chromosomes and help maintain genome integrity by protecting these vulnerable terminal ends from instability.
These telomeric regions are characterized by long regions of double-stranded CCCTAA:TTAGGG repeats. The repeats end with a 3’ protrusion of between 10 and 50 single-stranded TTAGGG repeats. The heterodimeric complex ribonucleoprotein enzyme telomerase adds TTAGGG repeats at the 3’ end of DNA strands. At these 3’ end protrusions, the G-rich overhang can form secondary structures such as G-quadraplexes if the overhang is longer than four TTAGGG repeats. The presence of these structures prevent telomere elongation by the telomerase complex.[16]
Telomeric quadruplexes
Telomeric repeats in a variety of organisms have been shown to form these quadruplex structures in vitro, and subsequently they have also been shown to form in vivo.[17][18] The human telomeric repeat (which is the same for all vertebrates) consists of many repeats of the sequenced (GGTTAG), and the quadruplexes formed by this structure have been well studied by NMR and X-ray crystal structure determination. The formation of these quadruplexes in telomeres has been shown to decrease the activity of the enzyme telomerase, which is responsible for maintaining length of telomeres and is involved in around 85% of all cancers. This is an active target of drug discovery, including telomestatin.
Non-telomeric quadruplexes
Quadruplexes are present in locations other than at the telomere. The proto-oncogene c-myc forms a quadruplex in a nuclease hypersensitive region critical for gene activity.[19][20] Other genes shown to form G-quadruplexes in their promoter regions include the chicken β-globin gene, human ubiquitin-ligase RFP2, and the proto-oncogenes c-kit, bcl-2, VEGF, H-ras and N-ras.[21][22][23]
Genome-wide surveys based on a quadruplex folding rule have been performed, which have identified 376,000 Putative Quadruplex Sequences (PQS) in the human genome, although not all of these probably form in vivo.[24] A similar study has identified putative G-quadruplexes in prokaryotes.[25] There are several possible models for how quadruplexes could influence gene activity, either by upregulation or downregulation. One model is shown below, with G-quadruplex formation in or near a promoter blocking transcription of the gene, and hence de-activating it. In another model, quadruplex formed at the non-coding DNA strand helps to maintain an open conformation of the coding DNA strand and enhance an expression of the respective gene.

G-Quadruplex function
It has been suggested that quadruplex formation plays a role in immunoglobulin heavy chain switching.[27] As cells have evolved mechanisms for resolving (i.e., unwinding) quadruplexes that form, quadruplex formation may be potentially damaging for a cell; the helicases WRN and Bloom syndrome protein have a high affinity for resolving DNA G-quadruplexes.[28] The DEAH/RHA helicase, DHX36, has also been identified as a key G-quadruplex resolvase.[29][30] More recently, there are many studies that implicate quadruplexes in both positive and negative transcriptional regulation, and in allowing programmed recombination of immunologlobin heavy genes and the pilin antigenic variation system of the pathogenic Neisseria.[31] The roles of quadruplex structure in translation control are not as well explored. The direct visualization of G-quadruplex structures in human cells[32] as well as the co-crystal structure of an RNA helicase bound to a G-quadruplex[33] have provided important confirmations of their relevance to cell biology. The potential positive and negative roles of quadruplexes in telomere replication and function remains controversial. T-loops and G-quadruplexes are described as the two tertiary DNA structures that protect telomere ends and regulate telomere length.[34]
Ligands which bind quadruplexes
One way of inducing or stabilizing G-quadruplex formation is to introduce a molecule which can bind to the G-quadruplex structure. A number of ligands, both small molecules and proteins, which can bind to the G-quadruplex. These ligands can be naturally occurring or synthetic. This has become an increasingly large field of research in genetics, biochemistry, and pharmacology.
A number of naturally occurring proteins have been identified which selectively bind to G-quadruplexes. These include the helicases implicated in Bloom's and Werner's syndromes and the Saccharomyces cerevisiae protein RAP1. In human organism, about 80 DNA or RNA quadruplex binding proteins were identified. [35] Recently, was found that all characterized G-quadruplex binding proteins share a 20 amino acid long motif/domain (RGRGR GRGGG SGGSG GRGRG) called NIQI (Novel Interesting Quadruplex Interaction Motif) [36]which is similar to the previously described RG-rich domain (RRGDG RRRGG GGRGQ GGRGR GGGFKG) of the FMR1 G-quadruplex binding protein [37] . An artificially derived three zinc finger protein called Gq1, which is specific for G-quadruplexes has also been developed, as have specific antibodies.
Cationic porphyrins have been shown to bind intercalatively with G-quadruplexes, as well as the molecule telomestatin.
Quadruplex prediction techniques
Identifying and predicting sequences which have the capacity to form quadruplexes is an important tool in further understanding their role. Generally, a simple pattern match is used for searching for possible intrastrand quadruplex forming sequences: d(G3+N1-7G3+N1-7G3+N1-7G3+), where N is any nucleotide base (including guanine).[38] This rule has been widely used in on-line algorithms. Although the rule effectively identifies sites of G-quadruplex formation it also identifies a subset of the imperfect homopurine mirror repeats capable of triplex formation[39] and C-strand i-motif formation.[40] Moreover, these sequences also have the capacity to form slipped and foldback structures that are implicit intermediates in the formation of both quadruplex[41] and triplex DNA[42] structures. In one study[43] it was found that the observed number per base pair (i.e. the frequency) of these motifs has increased rapidly in the eumetazoa for which complete genomic sequences are available. This suggests that the sequences may be under positive selection enabled by the evolution of systems capable of suppressing non-B structure formation.
Role in neurological disorders
G-quadruplexes have been implicated in neurological disorders through two main mechanisms. The first is through expansions of G-repeats within genes that lead to the formation of G-quadruplex structures that directly cause disease, as is the case with the C9orf72 gene and amyotrophic lateral sclerosis (ALS) or frontotemporal dementia (FTD). The second mechanism is through mutations that affect the expression of G-quadruplex binding proteins, as seen in the FMR1 gene and Fragile X Syndrome.[44]
The C9orf72 gene codes for the protein C9orf72 which is found throughout the brain in neuronal cytoplasm and at presynaptic terminals.[45] Mutations of the C9orf72 gene have been linked to the development of FTD and ALS.[46] These two diseases have a causal relationship to GGGGCC (G4C2) repeats within the 1st intron of C9orf72 gene. Normal individuals typically have around 2 to 8 G4C2 repeats, but individuals with FTD or ALS have from 500 to several thousand G4C2 repeats.[47][48] The transcribed RNA of these repeats have been shown to form stable G-quadruplexes, with evidence showing that the G4C2 repeats in DNA have the ability to form mixed parallel-antiparallel G-quadruplex structures as well.[49][50] These RNA transcripts containing G4C2 repeats were shown to bind and separate a wide variety of proteins, including nucleolin. Nucleolin is involved in the synthesis and maturation of ribosomes within the nucleus, and separation of nucleolin by the mutated RNA transcripts impairs nucleolar function and ribosomal RNA synthesis.[51]
Fragile X mental retardation protein (FMRP) is a widely expressed protein coded by the fragile X mental retardation gene 1 (FMR1) that binds to G-quadruplex secondary structures in neurons and is involved in synaptic plasticity.[52] FMRP acts as a negative regulator of translation, and its binding stabilizes G-quadruplex structures in mRNA transcripts, inhibiting ribosome elongation of mRNA in the neuron's dendrite and controlling the timing of the transcript's expression.[53][54] Mutations of this gene can cause the development of Fragile X Syndrome, autism, and other neurological disorders.[55] Specifically, Fragile X Syndrome is caused by an increase from 50 to over 200 CGG repeats within exon 13 of the FMR1 gene. This repeat expansion promotes DNA methylation and other epigenetic heterochromatin modifications of FMR1 that prevent the transcription of the gene, leading to pathological low levels of FMRP.[56][57]
Therapeutic approaches
Antisense-mediated interventions and small-molecule ligands are common strategies used to target neurological diseases linked to G-quadruplex expansion repeats. Therefore, these techniques are especially advantageous for targeting neurological diseases that have a gain-of-function mechanism, which is when the altered gene product has a new function or new expression of a gene; this has been detected in the C9orf72 (chromosome 9 open reading frame 72).[58]
Antisense therapy is the process by which synthesized strands of nucleic acids are used to bind directly and specifically to the mRNA produced by a certain gene, which will inactivate it. Antisense oligonucleotides (ASOs) are commonly used to target C9orf72 RNA of the G-quadruplex GGGGCC expansion repeat region, which has lowered the toxicity in cellular models of C9orf72.[59][60][61] ASOs have previously been used to restore normal phenotypes in other neurological diseases that have gain-of-function mechanisms, the only difference is that it was used in the absence of G-quadruplex expansion repeat regions.[62][63][64][65]
Another commonly used technique is the utilization of small-molecule ligands. These can be used to target G-quadruplex regions that cause neurological disorders. Approximately 1,000 various G-quadruplex ligands exist in which they are able to interact via their aromatic rings; this allows the small-molecule ligands to stack on the planar terminal tetrads within the G-quadruplex regions. A disadvantage of using small-molecule ligands as a therapeutic technique is that specificity is difficult to manage due to the variability of G-quadruplexes in their primary sequences, orientation, thermodynamic stability, and nucleic acid strand stoichiometry. As of now, no single small-molecule ligand has been able to be 100% specific for a single G-quadruplex sequence.[66][67] However, a cationic porphyrin known as TMPyP4 is able to bind to the C9orf72 GGGGCC repeat region, which causes the G-quadruplex repeat region to unfold and lose its interactions with proteins causing it to lose its functionality.[68] Small-molecule ligands, composed primarily of lead, can target GGGGCC repeat regions as well and ultimately decreased both repeat-associated non-ATG translation and RNA foci in neuron cells derived from patients with Amyotrophic lateral sclerosis (ALS). This provides evidence that small-molecule ligands are an effective and efficient process to target GGGGCC regions, and that specificity for small-molecule ligand binding is a feasible goal for the scientific community.
Metal complexes have a number of features that make them particularly suitable as G4 DNA binders and therefore as potential drugs. While the metal plays largely a structural role in most G4 binders, there are also examples where it interacts directly with G4s by electrostatic interactions or direct coordination with nucleobases.[69]
Notes
- ↑ Routh, Eric (2017). "A G-quadruplex DNA-affinity approach for purification of enzymatically active G4 Resolvase1". J. Vis. Exp. 121 (121). doi:10.3791/55496. PMID 28362374.
- 1 2 Largy, Eric; Mergny, Jean-Louis; Gabelica, Valérie (2016). "Chapter 7. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability". In Astrid, Sigel; Helmut, Sigel; Roland K.O., Sigel. The Alkali Metal Ions: Their Role in Life. Metal Ions in Life Sciences. 16. Springer. pp. 203–258. doi:10.1007/978-4-319-21756-7_7 (inactive 2018-09-18).
- ↑ Sundquist, Wesley; Klug, Aaron (1989). "Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops". Nature. 342 (6251): 825–829. doi:10.1038/342825a0. PMID 2601741.
- 1 2 Sen, Dipankar; Gilbert, Walter (1988). "Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis". Nature. 334 (6180): 364–366. doi:10.1038/334364a0. PMID 3393228.
- ↑ Han, Haiyong (2000). "G-quadruplex DNA: a potential target for anti-cancer drug design". TiPS. 21 (4): 136–142. doi:10.1016/s0165-6147(00)01457-7.
- ↑ Bochman, Matthew L.; Paeschke, Katrin; Zakian, Virginia A. (2012). "DNA secondary structures: stability and function of G-quadruplex structures". Nature Reviews. Genetics. 13 (11): 770–780. doi:10.1038/nrg3296. PMC 3725559. PMID 23032257.
- ↑ Rhodes, Daniela; Lipps, Hans J. (2015). "G-quadruplexes and their regulatory roles in biology". Nucleic Acids Res. 43 (18): 8627–8637. doi:10.1093/nar/gkv862. PMC 4605312. PMID 26350216.
- ↑ Simonsson, T. (2001). "G-Quadruplex DNA Structures Variations on a Theme". Biological Chemistry. 382 (4): 621–628. doi:10.1515/BC.2001.073. PMID 11405224.
- ↑ Burge, S.; Parkinson, G. N.; Hazel, P.; Todd, A. K.; Neidle, S. (2006). "Quadruplex DNA: Sequence, topology and structure". Nucleic Acids Research. 34 (19): 5402–5415. doi:10.1093/nar/gkl655. PMC 1636468. PMID 17012276.
- ↑ Cao, K.; Ryvkin, P.; Johnson, FB. (2012). "Computational detection and analysis of sequences with duplex-derived interstrand G-quadruplex forming potential". Methods. 57 (1): 3–10. doi:10.1016/j.ymeth.2012.05.002. PMC 3701776. PMID 22652626.
- ↑ Kudlicki, A. (2016). "G-Quadruplexes Involving Both Strands of Genomic DNA Are Highly Abundant and Colocalize with Functional Sites in the Human Genome". PLoS ONE. 11 (1): e0146174. doi:10.1371/journal.pone.0146174. PMC 4699641. PMID 26727593.
- ↑ Murat, P.; Balasubramanian, S. (2014). "Existence and consequences of G-quadruplex structures in DNA". Curr. Opin. Genet. Dev. 25 (25): 22–29. doi:10.1016/j.gde.2013.10.012. PMID 24584093.
- ↑ Miyoshi, Daisuke; Karimata, Hisae; Sugimoto, Naoki (May 27, 2006). "Hydration Regulates Thermodynamics of G-Quadruplex Formation under Molecular Crowding Conditions". Journal of the American Chemical Society. 128 (24): 7957–7963. doi:10.1021/ja061267m. PMID 16771510.
- ↑ Zheng, KW; Chen, Z; Hao, YH; Tan, Z (January 2010). "Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA". Nucleic Acids Res. 38 (1): 327–338. doi:10.1093/nar/gkp898. PMC 2800236. PMID 19858105.
- ↑ Tamaki, Endoh; Rode, Ambadas B.; Takahashi, Shuntaro; Kataoka, Yuka; Kuwahara, Masayasu; Sugimoto, Naoki (January 2016). "Real-Time Monitoring of G-Quadruplex Formation during Transcription". Analytical Chemistry. 88 (4): 1984–1989. doi:10.1021/acs.analchem.5b04396. PMID 26810457.
- ↑ Wang, Q; Liu, J.Q.; Chen, Z; Zheng, K.W.; Chen, C.Y.; Hao, Y.H.; Tan, Z (2011). "G-quadruplex formation at the 3' end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase". Nucleic Acids Res. 39 (39): 6229–6237. doi:10.1093/nar/gkr164. PMC 3152327. PMID 21441540.
- ↑ Schaffitzel, C; Berger, I; Postberg, J; Hanes, J; Lipps, H. J.; Plückthun, A (2001). "In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei". Proceedings of the National Academy of Sciences. 98 (15): 8572–7. doi:10.1073/pnas.141229498. PMC 37477. PMID 11438689.
- ↑ Paeschke, K.; Simonsson, T.; Postberg, J.; Rhodes, D.; Lipps, H. J. (2005). "Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo". Nature Structural & Molecular Biology. 12 (10): 847–854. doi:10.1038/nsmb982. PMID 16142245.
- ↑ Simonsson, T.; Pecinka, P.; Kubista, M. (1998). "DNA tetraplex formation in the control region of c-myc". Nucleic Acids Research. 26 (5): 1167–1172. doi:10.1093/nar/26.5.1167. PMC 147388. PMID 9469822.
- ↑ Siddiqui-Jain, A.; Grand, C. L.; Bearss, D. J.; Hurley, L. H. (2002). "Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription". Proceedings of the National Academy of Sciences. 99 (18): 11593–11598. doi:10.1073/pnas.182256799. PMC 129314. PMID 12195017.
- ↑ Huppert, Julian L.; Balasubramanian, Shankar (14 December 2006). "G-quadruplexes in promoters throughout the human genome". Nucleic Acids Research. 35 (2): 406–413. doi:10.1093/nar/gkl1057. PMC 1802602. PMID 17169996.
- ↑ Dai, Jixun; et al. (5 January 2006). "An Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel G-Strands Formed in the Human BCL-2 Promoter Region in Solution". J. Am. Chem. Soc. 128 (4): 1096–1098. doi:10.1021/ja055636a. PMC 2556172. PMID 16433524.
- ↑ Fernando, Himesh; et al. (28 May 2006). "A Conserved Quadruplex Motif Located in a Transcription Activation Site of the Human c-kit Oncogene". Biochemistry. 45 (25): 7854–7860. doi:10.1021/bi0601510. PMC 2195898. PMID 16784237.
- ↑ Huppert, J. L.; Balasubramanian, S. (2005). "Prevalence of quadruplexes in the human genome". Nucleic Acids Research. 33 (9): 2908–2916. doi:10.1093/nar/gki609. PMC 1140081. PMID 15914667.
- ↑ Rawal, P.; Kummarasetti, V. B.; Ravindran, J.; Kumar, N.; Halder, K.; Sharma, R.; Mukerji, M.; Das, S. K.; Chowdhury, S. (2006). "Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation". Genome Research. 16 (5): 644–655. doi:10.1101/gr.4508806. PMC 1457047. PMID 16651665.
- ↑ Bugaut A, Balasubramanian S (2012). "5'-UTR RNA G-quadruplexes: translation regulation and targeting". Nucleic Acids Res. 40 (11): 4727–41. doi:10.1093/nar/gks068. PMC 3367173. PMID 22351747.
- ↑ Sen, D.; Gilbert, W. (1988). "Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis". Nature. 334 (6180): 364–366. doi:10.1038/334364a0. PMID 3393228.
- ↑ Kamath-Loeb, A.; Loeb, L. A.; Fry, M. (2012). Cotterill, Sue, ed. "The Werner Syndrome Protein is Distinguished from the Bloom Syndrome Protein by Its Capacity to Tightly Bind Diverse DNA Structures". PLoS ONE. 7 (1): e30189. doi:10.1371/journal.pone.0030189. PMC 3260238. PMID 22272300.
- ↑ Vaughn, James P.; Creacy, Steven D.; Routh, Eric D.; Joyner-Butt, Christi; Jenkins, G. Scott; Pauli, Sandra; Nagamine, Yoshikuni; Akman, Steven A. (18 November 2005). "The DEXH Protein Product of the DHX36 Gene Is the Major Source of Tetramolecular Quadruplex G4-DNA Resolving Activity in HeLa Cell Lysates". Journal of Biological Chemistry. 280 (46): 38117–38120. doi:10.1074/jbc.C500348200. PMID 16150737.
- ↑ Chen, Michael C.; Ferré-D’Amaré, Adrian R. (15 August 2017). "Structural Basis of DEAH/RHA Helicase Activity". Crystals. 7 (8): 253. doi:10.3390/cryst7080253.
- ↑ Maizels, N.; Gray, L. T. (2013). Rosenberg, Susan M, ed. "The G4 Genome". PLoS Genetics. 9 (4): e1003468. doi:10.1371/journal.pgen.1003468. PMC 3630100. PMID 23637633.
- ↑ Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. (2013). "Quantitative visualization of DNA G-quadruplex structures in human cells". Nature Chemistry. 5 (3): 182–186. doi:10.1038/nchem.1548. PMC 3622242. PMID 23422559.
- ↑ Chen, Michael C.; Tippana, Ramreddy; Demeshkina, Natalia A.; Murat, Pierre; Balasubramanian, Shankar; Myong, Sua; Ferré-D’Amaré, Adrian R. (June 2018). "Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36". Nature. 558 (7710): 465–469. doi:10.1038/s41586-018-0209-9. ISSN 0028-0836. PMID 29899445.
- ↑ Rice C, Skordalakes E (2016). "Structure and function of the telomeric CST complex". Computational and Structural Biotechnology Journal. 14: 161–167. doi:10.1016/j.csbj.2016.04.002. PMC 4872678. PMID 27239262.
- ↑ Brázda V, Hároníková L, Liao JC, Fojta M (2014). "DNA and RNA G-quadruplex-binding proteins". International Journal of Molecular Sciences . 15 (10): 17493–17517. doi:10.3390/ijms151017493. PMC 4227175. PMID 25268620.
- ↑ Brázda V, Červeň J, Bartas M, Mikysková N, Coufal J, Pečinka P (2018). "The Amino Acid Composition of Quadruplex Binding Proteins Reveals a Shared Motif and Predicts New Potential Quadruplex Interactors". Molecules. 23 (9): E2341. doi:10.3390/molecules23092341. PMID 30216987.
- ↑ Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, Darnell RB, Patel DJ (2011). "Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction". Nature Structural & Molecular Biology. 18 (7): 796–804. doi:10.1038/nsmb.2064. PMC 3130835. PMID 21642970.
- ↑ Todd, A. K.; Johnston, M.; Neidle, S. (2005). "Highly prevalent putative quadruplex sequence motifs in human DNA". Nucleic Acids Research. 33 (9): 2901–2907. doi:10.1093/nar/gki553. PMC 1140077. PMID 15914666.
- ↑ Frank-Kamenetskii, M. D.; Mirkin, S. M (1995). "Triplex DNA structures". Annual Review of Biochemistry. 64 (9): 65–95. doi:10.1146/annurev.bi.64.070195.000433. PMID 7574496.
- ↑ Guo, K.; Gokhale, V.; Hurley, L. H.; Sun, D (2008). "Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene". Nucleic Acids Research. 36 (14): 4598–4608. doi:10.1093/nar/gkn380. PMC 2504309. PMID 18614607.
- ↑ Sundquist, W. I.; Klug, M. (1989). "Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops". Nature. 342 (6251): 825–829. doi:10.1038/342825a0. PMID 2601741.
- ↑ Mirkin, S. M.; Lyamichev, V. I.; Drushlyak, K. V.; Dobrynin, V. N.; Filippov, S. A.; Frank-Kamenetskii, M. D. (1987). "DNA H form requires a homopurine-homopyrimidine mirror repeat". Nature. 330 (6147): 495–497. doi:10.1038/330495a0. PMID 2825028.
- ↑ Smith, S. S. (2010). "Evolutionary expansion of structurally complex DNA sequences". Cancer Genomics and Proteomics. 7 (4): 207–216. PMID 20656986.
- ↑ Roberto Simone, Pietro Fratta, Stephen Neidle, Gary N. Parkinson, Adrian M. Isaacs (May 2015). "G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome". Federation of European Biochemical Societies. 589 (14): 1653–1668. doi:10.1016/j.febslet.2015.05.003. PMID 25979174.
- ↑ C9orf72 chromosome 9 open reading frame 72 [Homo sapiens] - Gene - NCBI
- ↑ Ratnavalli E, Brayne C, Dawson K, Hodges JR (2002). "The prevalence of frontotemporal dementia". Neurology. 58 (11): 1615–21. doi:10.1212/WNL.58.11.1615. PMID 12058088.
- ↑ Rutherford NJ, Heckman MG, Dejesus-Hernandez M, Baker MC, Soto-Ortolaza AI, Rayaprolu S, Stewart H, Finger E, Volkening K, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio EH, Lippa C, Knopman DS, Kretzschmar HA, Neumann M, Caselli RJ, White CL 3rd, Mackenzie IR, Petersen RC, Strong MJ, Miller BL, Boeve BF, Uitti RJ, Boylan KB, Wszolek ZK, Graff-Radford NR, Dickson DW, Ross OA, Rademakers R. (Dec 2012). "Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype". Neurobiology of Aging. 33 (12): 2950.e5–7. doi:10.1016/j.neurobiolaging.2012.07.005. PMC 3617405. PMID 22840558.
- ↑ Jon Beck, Mark Poulter, Davina Hensman, Jonathan D. Rohrer, Colin J. Mahoney, Gary Adamson, Tracy Campbell, James Uphill, Aaron Borg, Pietro Fratta, Richard W. Orrell, Andrea Malaspina, James Rowe, Jeremy Brown, John Hodges, Katie Sidle, James M. Polke, Henry Houlden, Jonathan M. Schott, Nick C. Fox, Martin N. Rossor, Sarah J. Tabrizi, Adrian M. Isaacs, John Hardy, Jason D. Warren, John Collinge, and Simon Mead (March 2013). "Large C9orf72 Hexanucleotide Repeat Expansions Are Seen in Multiple Neurodegenerative Syndromes and Are More Frequent Than Expected in the UK Population". The American Journal of Human Genetics. 92 (3): 345–353. doi:10.1016/j.ajhg.2013.01.011. PMC 3591848. PMID 23434116.
- ↑ Pietro Fratta, Sarah Mizielinska, Andrew J. Nicoll, Mire Zloh, Elizabeth M. C. Fisher, Gary Parkinson, and Adrian M. Isaacs (December 2012). "C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes". Scientific Reports. 2: 1016. doi:10.1038/srep01016. PMC 3527825. PMID 23264878.
- ↑ Kaalak Reddy, Bita Zamiri, Sabrina Y. R. Stanley, Robert B. Macgregor Jr. and Christopher E. Pearson (April 2013). "The Disease-associated r(GGGGCC)n Repeat from the C9orf72 Gene Forms Tract Length-dependent Uni- and Multimolecular RNA G-quadruplex Structures". Journal of Biological Chemistry. 288 (14): 9860–9866. doi:10.1074/jbc.C113.452532. PMC 3617286. PMID 23423380.
- ↑ Aaron R. Haeusler, Christopher J. Donnelly, Goran Periz, Eric A. J. Simko, Patrick G. Shaw, Min-Sik Kim, Nicholas J. Maragakis, Juan C. Troncoso, Akhilesh Pandey, Rita Sattler, Jeffrey D. Rothstein & Jiou Wang (March 2014). "C9orf72 nucleotide repeat structures initiate molecular cascades of disease". Nature. 507 (7491): 195–200. doi:10.1038/nature13124. PMC 4046618. PMID 24598541.
- ↑ Darnell, J. C., Jensen, K. B., Jin, P., Brown, V., Warren, S. T., Darnell. R. B. (November 2001). "Fragile X Mental Retardation Protein Targets G Quartet mRNAs Important for Neuronal Function". Cell. 107 (4): 489–499. doi:10.1016/S0092-8674(01)00566-9.
- ↑ Ceman, S., O'Donnell, W. T., Reed, M., Patton, S., Pohl, J., Warren, S. T. (December 2003). "Phosphorylation influences the translation state of FMRP-associated polyribosomes". Human Molecular Genetics. 12 (24): 3295–3305. doi:10.1093/hmg/ddg350. PMID 14570712.
- ↑ Fahling, M., Mrowka, R., Steege, A., Kirschner, K. M., Benko, E., Forstera, B., Persson, P. B., Thiele, B. J., Meier, J. C., Scholz, H. (Dec 2008). "Translational Regulation of the Human Achaete-scute Homologue-1 by Fragile X Mental Retardation Protein". Journal of Biological Chemistry. 284 (7): 4255–4266. doi:10.1074/jbc.M807354200. PMID 19097999.
- ↑ "Fragile X Mental Retardation" The Human Gene Compendium
- ↑ Pieretti, M., Zhang, F., Fu, Y., Warren, S. T., Oostra, B. A., Caskey, C. T., Nelson, D. L. (August 1991). "Absence of expression of the FMR-1 gene in fragile X syndrome". Cell. 66 (4): 816–822. doi:10.1016/0092-8674(91)90125-I.
- ↑ Sutcliffe, J. S., Nelson, D. L., Zhang, F., Pieretti, M., Caskey, C. T., Saxe, D., Warren, S. T. (September 1992). "DNA methylation represses FMR-1 transcription in fragile X syndrome". Human Molecular Genetics. 1 (6): 397–400. doi:10.1093/hmg/1.6.397. PMID 1301913.
- ↑ {{Mizielinska, S., Isaacs, A.M. (2014). C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia: gain or loss of function? Curr. Opin. Neurol. 27. 515-523.}}
- ↑ Donnelly, C.J., Zhang, P.W., et. al. (2013). RNA toxicity from the ALS/FTD C9orf72 expansion is mitigated by antisense intervention. Neuron. 80. 415-428.
- ↑ Lagier-Tourenne, C., Baughn, M., et. al. (2013). Targeted degradation of sense and antisense C9orf72 foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U.S.A. 110. 4530-4539.
- ↑ Sareen, D., O’Rourke, J.G., et. al. (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with C9orf72 repeat expansion. Sci. Transl. Med. 5. 208ra149.
- ↑ Wheeler, T.M., Leger, A.J., et. al. (2012). Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 488. 111-115.
- ↑ Lee, J.E., Bennett, C.F., and Cooper, T.A. (2012). RNAse H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc. Natl. Acad. Sci. U.S.A. 109. 4221-4226.
- ↑ Carroll, J.B., Warby., S.C., et. al. (2011). Potent and selective antisense oligonucleotides targeting single nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant Huntingtin. Mol. Ther. 19. 2178-2185.
- ↑ Gagnen, K.T., Pendergraff, H.M. (2010). Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochem. 49. 10166-10178.
- ↑ Campbell, N.H., Patel, M., et al. (2009). Selective in ligand recognition of G-quadruplex loops. Biochem. 48. 1675-1680.
- ↑ Ohnmacht, S.A., and Neidle, S. (2014). Small-molecule quadruplex targeted drug discovery. Bioorg. Med. Chem. Lett. 24. 2602-2612.
- ↑ Zamiri, B., Reddy, K., et. al. (2014). TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interactions of RNA-binding proteins. J. Biol. Chem. 289. 4653-4659.
- ↑ Vilar, Ramon (2018). "Chapter 12. Nucleic Acid Quadruplexes and Metallo-Drugs". In Sigel, Astrid; Sigel, Helmut; Freisinger, Eva; Sigel, Roland K. O. Metallo-Drugs: Development and Action of Anticancer Agents. Metal Ions in Life Sciences. 18. pp. 325–349. doi:10.1515/9783110470734-018. ISBN 9783110470734. PMID 29394031.
References
- Jiangtao Ren; Jiahai Wang*; Lei Han; Erkang Wang*; Jin Wang* (2011). "Kinetically grafting G-qaudruplex onto DNA nanostructure as structure and function encoding via DNA machine". Chem. Commun. 47 (38): 10563–10565. doi:10.1039/c1cc13973h. PMID 21858307.
- Johnson JE, Smith JS, Kozak ML, Johnson FB (2008). "In vivo veritas: using yeast to probe the biological functions of G-quadruplexes". Biochimie. 90 (8): 1250–1263. doi:10.1016/j.biochi.2008.02.013. PMC 2585026. PMID 18331848.
- Huppert JL, Balasubramanian S (2005). "Prevalence of quadruplexes in the human genome". Nucleic Acids Research. 33 (9): 2908–2916. doi:10.1093/nar/gki609. PMC 1140081. PMID 15914667.
- Todd AK, Johnston M, Neidle S (2005). "Highly prevalent putative quadruplex sequence motifs in human DNA". Nucleic Acids Research. 33 (9): 2901–2907. doi:10.1093/nar/gki553. PMC 1140077. PMID 15914666.
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006). "Quadruplex DNA: sequence, topology and structure". Nucleic Acids Research. 34 (19): 5402–5415. doi:10.1093/nar/gkl655. PMC 1636468. PMID 17012276.
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH (2002). "Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription". PNAS. 99 (18): 11593–8. doi:10.1073/pnas.182256799. PMC 129314. PMID 12195017.
- Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S (2006). "Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation". Genome Res. 16 (5): 644–55. doi:10.1101/gr.4508806. PMC 1457047. PMID 16651665.
- Xu Hou; Wei Guo; Fan Xia; Fu-Qiang Nie; Hua Dong; Ye Tian; Liping Wen; Lin Wang; Liuxuan Cao; Yang Yang; Jianming Xue; Yanlin Song; Yugang Wang; Dongsheng Liu; Lei Jiang (2009). "A biomimetic potassium responsive nanochannel: G-quadruplex DNA conformational switching in a synthetic nanopore". J. Am. Chem. Soc. 131 (22): 7800–7805. doi:10.1021/ja901574c. PMID 19435350.
- Neidle & Balasubramanian, ed. (2006). Quadruplex Nucleic Acids. ISBN 978-0-85404-374-3. Archived from the original on 2007-09-30.
- Rowland, Gerald B.; Barnett, Kerry; DuPont, Jesse I.; Akurathi, Gopalakrishna; Le, Vu H.; Lewis, Edwin A (1 December 2013). "The effect of pyridyl substituents on the thermodynamics of porphyrin binding to G-quadruplex DNA". Bioorganic & Medicinal Chemistry. 21 (23): 7515–22. doi:10.1016/j.bmc.2013.09.036. PMID 24148836.
External links
- Nanopore and Aptamer Biosensor group{NAB group}
Quadruplex websites
- G-Quadruplex World – a website to discuss publications and other information of interest to those working in the field of G-quadruplexes
- Quadbase – downloadable data on predicted G-quadruplexes
- Greglist – a database listing potential G-quadruplex regulated genes
- Database on Quadruplex information: QuadBase from IGIB
- GRSDB- a database of G-quadruplexes near RNA processing sites.
- GRS_UTRdb- a database of G-quadruplexes in the UTRs.
- G-quadruplex Resource Site
- non-B Motif Search Tool at non-B DB- a web server to predict G-quadruplex forming motifs and other non-B DNA forming motifs from users' DNA sequences.
Tools to predict G-quadruplex motifs
- Quadparser: Downloadable program for finding putative quadruplex-forming sequences from Balasubramanian's group.
- QGRS Mapper: a web-based application for predicting G-quadruplexes in nucleotide sequences and NCBI genes from Bagga's group.
- Quadfinder: Tool for Prediction and Analysis of G Quadruplex Motifs in DNA/RNA Sequences from Maiti's group, IGIB, Delhi, India
- G4Hunter from Mergny's group but user need to run the code in R.
- pqsfinder: an exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R.