Clostridioides difficile (bacteria)
| Clostridioides difficile | |
|---|---|
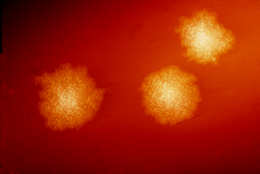 | |
| C. difficile colonies on a blood agar plate | |
 | |
| Micrograph of Clostridioides difficile | |
| Scientific classification | |
| Kingdom: | Bacteria |
| Phylum: | Firmicutes |
| Class: | Clostridia |
| Order: | Clostridiales |
| Family: | Peptostreptococcaceae |
| Genus: | Clostridioides |
| Species: | C. difficile |
| Binomial name | |
| Clostridioides difficile Hall & O'Toole, 1935; Lawson & Rainey, 2016 | |
| Synonyms | |
|
Clostridium difficile | |
Clostridioides difficile (syn. Clostridium difficile), also known as C. difficile, C. diff (/siː
Clostridioides are anaerobic, motile bacteria, ubiquitous in nature, and especially prevalent in soil. Its vegetative cells are rod shaped, pleomorphic, and occur in pairs or short chains. Under the microscope, they appear as long, irregular (often drumstick- or spindle-shaped) cells with a bulge at their terminal ends (forms subterminal spores). Under Gram staining, C. difficile cells are Gram-positive and show optimum growth on blood agar at human body temperatures in the absence of oxygen. C. difficile is catalase and superoxide dismutase negative, and produces two types of toxins: enterotoxin A and cytotoxin B, which disrupts cytoskeleton signal transductions in the host. When stressed, the bacteria produce spores that are able to tolerate extreme conditions that the active bacteria cannot tolerate.[2]
C. difficile may become established in the human colon; it is present in 2–5% of the adult population.[2] Sometimes antibiotic therapy for various infections has the adverse effect of disrupting the normal balance of the gut microbiota, in which case C. difficile may opportunistically dominate, causing C. difficile infection.
Taxonomy
This species has been officially renamed, in 2016, from Clostridium difficile to Clostridioides difficile.[3][4] This new name reflects the taxonomic differences between this species and other members of the Clostridium genus, while maintaining the common name as C. diff.[5] Currently, the only other species in this new Clostridioides genus is Clostridioides mangenotii (formerly known as Clostridium mangenotii).[6] It had previously been proposed, in a July 2013 paper from Environmental Microbiology, to rename the species Peptoclostridium difficile.[7][8]
The species name is New Latin, from the Greek kloster (κλωστήρ), "spindle",[9] and Latin difficile, "difficult, obstinate".[10] It is pronounced /klɔːˈstrɪdiəm
Human pathogen
Pathogenic C. difficile strains produce multiple toxins.[12] The best-characterized are enterotoxin (C. difficile toxin A) and cytotoxin (C. difficile toxin B), both of which may produce diarrhea and inflammation in infected patients (C. difficile colitis), although their relative contributions have been debated. The diarrhea may range from a few days of intestinal fluid loss to life-threatening pseudomembranous colitis. Pseudomembranous colitis is associated with intense inflammation of the colon and formation of pseudomembranes on the intestinal mucosal surface.[2] Toxins A and B are glucosyltransferases that target and inactivate the Rho family of GTPases. Toxin B (cytotoxin) induces actin depolymerization by a mechanism correlated with a decrease in the ADP-ribosylation of the low molecular mass GTP-binding Rho proteins.[13] Another toxin, binary toxin, also has been described, but its role in disease is not fully understood.[14]
Additional virulence factors include an adhesin factor which mediate the binding to human colonic cells and a hyaluronidase.[15]
Antibiotic treatment of C. diff infections may be difficult, due both to antibiotic resistance and physiological factors of the bacterium (spore formation, protective effects of the pseudomembrane).[2] The emergence of a new, highly toxic strain of C. difficile, resistant to fluoroquinolone antibiotics, such as ciprofloxacin and levofloxacin, said to be causing geographically dispersed outbreaks in North America, was reported in 2005.[16] The U.S. Centers for Disease Control (CDC) in Atlanta warned of the emergence of an epidemic strain with increased virulence, antibiotic resistance, or both.[17]
Transmission
Clostridioides difficile is transmitted from person to person by the fecal-oral route. C. difficile is shed in faeces. Any surface, device, or material (e.g., toilets, bathing tubs, and electronic rectal thermometers) that becomes contaminated with faeces may serve as a reservoir for the C. difficile spores. C. difficile spores are transferred to patients mainly via the hands of healthcare personnel who have touched a contaminated surface or item. C. difficile can live for long periods of time on surfaces.[18] The organism forms heat-resistant spores that are not killed by alcohol-based hand cleansers or routine surface cleaning, thus, these spores survive in clinical environments for long periods. Because of this, the bacterium may be cultured from almost any surface. Once spores are ingested, their acid-resistance allows them to pass through the stomach unscathed. They germinate and multiply into vegetative cells in the colon upon exposure to bile acids. Consequently, the World Health Organization advocates the use of soap in addition to alcohol solutions in order to limit the spread of the spores.[19]
Host range
C. difficile infects pigs, calves, and humans, and inhabits a natural reservoir of soil, faeces of domestic animals and humans, sewage, the human intestinal tract, and retail meat.[20]
A 2015 CDC study estimated that C. diff afflicted almost half a million Americans and caused 29,000 deaths in 2011. The study estimated that 40 percent of cases began in nursing homes or community health care settings, while 24 percent occurred in hospitals.[21]
C. difficile is common in the human digestive system. However, C. difficile is a poor competitor, and is often out competed by other bacteria for nutrients in the digestive system. As a result, C. difficile is kept to a manageable amount. If the sudden introduction of antibiotic disrupts the microbiome, C. difficile may be able to grow as a result of many of its competitors being killed off. The incubation period is 5–10 days, with a range of 1 day to weeks following antibiotic treatment for AAD (antibiotic associated diarrhea). Additionally, carriage of C. difficile with high levels of toxins are common in young children while disease is rare. The production of one or even both toxins is not always sufficient for producing symptoms.[22]
Signs and symptoms
Symptoms of C. difficile include: Diarrhea[23]—watery, at least a minimum of three times a day and up to 15 times a day. Abdominal cramping and pain that can be severe. Loss of appetite and dehydration. Weight loss and nausea.[24]
Treatment
Patients being treated with antibiotics when symptoms begin should stop taking them, if possible. This break in antibiotic therapy can sometimes lead to spontaneous resolution of symptoms. Patients that don't respond to the cessation of broad-spectrum antibiotics will need to be treated with antibiotics capable of killing C. difficile spores. Primary infections are typically treated with metronidazole. This medication is normally given 3 times a day orally and should be taken for a minimum of 10 days. Some patients can't tolerate the side effects of metronidazole, which include severe nausea and vomiting, which can lead to dehydration. In this case, patients should be given oral vancomycin, usually at a dosage of 125 milligrams every 6 hours.[25]
About 20% of patients who successfully complete therapy of primary infection with metronidazole or vancomycin will experience a relapse. A fraction of those patients will experience continuous reoccurrences of the infection. The first relapse of a C. difficile is usually treated with the same antibiotic used to treat the primary infection. Any subsequent infections should not be treated with metronidazole. Occasionally, a standard 10 day course of oral vancomycin will not work. In these cases, a vancomycin taper is the preferred treatment. Patients will take decreasing doses of vancomycin over a period of up to 3 months, depending on the severity of the infection.[24]
Each subsequent relapse of C. difficile tends to be more severe than previous infections. Long term treatment with a vancomycin taper supplemented with probiotics, especially Saccharomyces boulardii, is associated with a higher rate of success.[26]
After three relapses, patients may be treated with oral fidaxomicin, a narrow spectrum antibiotic. The usual dosage is 200 mg twice a day orally for 10 days. Fidaxomicin is considered to be superior to vancomycin for severe CDI (C. difficile infection).[25] The major downside of treatment with fidaxomicin is the cost of medication. A 10-day course may cost up to 3500 USD.
Patients that do not respond to traditional antibiotic therapy may be eligible for a fecal transplant. Healthcare providers can transfer stool from a healthy person to the colon of a patient with repeated CDI. This process is the most successful treatment for severe CDI with a cure rate of around 93%. Long term effects of fecal transplantation are unknown, as the procedure has only been FDA approved since 2011 and relatively few procedures have been performed. If transplantation is not an option, removal of the infected part of the colon can cure C. difficile.[25][24]
Strains
In 2005, molecular analysis led to the identification of the C. difficile strain type characterized as group BI by restriction endonuclease analysis, as North American pulse-field-type NAP1 by pulsed-field gel electrophoresis and as ribotype 027; the differing terminology reflects the predominant techniques used for epidemiological typing. This strain is referred to as C. difficile BI/NAP1/027.[27]
Two strains, ribotypes RT078 and RT027, can live on low concentrations of the sugar trehalose; both strains became more common after trehalose was introduced as a food additive in the early 2000s and thus increasing dietary trehalose intake.[28]
Genome
| NCBI genome ID | 535? |
|---|---|
| Ploidy | haploid |
| Genome size | 4.3 Mb |
| Number of chromosomes | 1 |
| Year of completion | 2005 |
The first complete genome sequence of a C. difficile strain was first published in 2005 by Sanger Institute in the UK. This was of the strain 630, a virulent and multiple drug-resistant strain isolated in Switzerland in 1982. Scientists at Sanger Institute have sequenced genomes of about 30 C. difficile isolates using next-generation sequencing technologies from 454 Life Sciences and Illumina.[29]
Researchers at McGill University in Montreal sequenced the genome of the highly virulent Quebec strain of C. difficile in 2005 using ultra-high-throughput sequencing technology. The tests involved doing 400,000 DNA parallel-sequencing reactions of the bacterium's genome, which had been fragmented for sequencing. These sequences were assembled computationally to form a complete genome sequence.[16][30]
In 2012, scientists at University of Oxford sequenced C. difficile genomes from 486 cases arising over four years in Oxfordshire using next-generation sequencing technologies from Illumina.[31]
Bacteriophage
At least eight mainly temperate bacteriophages have been isolated from C. difficile, ranging in genome size from about 30 to about 60 kb.[32] Both environmentally and clinically derived C. difficile strains carry a diverse and prevalent set of prophages.[32]
Notes
- ↑ Moreno MA, Furtner F, Rivara FP (June 2013). "Clostridium difficile: A Cause of Diarrhea in Children". JAMA Pediatrics. 167 (6): 592. doi:10.1001/jamapediatrics.2013.2551. PMID 23733223.
- 1 2 3 4 Ryan KJ, Ray CG, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. pp. 322–4. ISBN 0-8385-8529-9.
- ↑ Oren, Aharon; Garrity, George M. (2017). "List of new names and new combinations previously effectively, but not validly, published". International Journal of Systematic and Evolutionary Microbiology. 67 (9): 3140–3143. doi:10.1099/ijsem.0.002278. PMC 5817221. PMID 28891789.
- ↑ Zhu, Duolong; Sorg, Joseph A.; Sun, Xingmin (2018). "Clostridioides difficile Biology: Sporulation, Germination, and Corresponding Therapies for C. difficile Infection". Frontiers in Cellular and Infection Microbiology. 8. doi:10.3389/fcimb.2018.00029. ISSN 2235-2988. PMC 5809512. PMID 29473021.
- ↑ Lawson, Paul A.; Citron, Diane M.; Tyrrell, Kerin L.; Finegold, Sydney M. (August 2016). "Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prévot 1938". Anaerobe. 40: 95–99. doi:10.1016/j.anaerobe.2016.06.008. ISSN 1075-9964.
- ↑ Galperin, Michael Y.; Brover, Vyacheslav; Tolstoy, Igor; Yutin, Natalya (2016). "Phylogenomic analysis of the family Peptostreptococcaceae (Clostridium cluster XI) and proposal for reclassification of Clostridium litorale (Fendrich et al. 1991) and Eubacterium acidaminophilum (Zindel et al. 1989) as Peptoclostridium litorale gen. nov. comb. nov. and Peptoclostridium acidaminophilum comb. nov". International Journal of Systematic and Evolutionary Microbiology. 66 (12): 5506–5513. doi:10.1099/ijsem.0.001548. PMC 5244501. PMID 27902180.
- ↑ Yutin N, Galperin MY (2013). "A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia". Environ. Microbiol. 15 (10): 2631–41. doi:10.1111/1462-2920.12173. PMC 4056668. PMID 23834245.
- ↑ Gerard J. Tortora; Berdell R. Funke; Christine L. Case (2015-01-13). Microbiology: An Introduction. Pearson Education. ISBN 978-0-13-392339-1.
- ↑ Liddell-Scott. "κλωστήρ". Greek-English Lexicon. Oxford{{inconsistent citations}}
- ↑ Cawley, Kevin. "Difficilis". Latin Dictionary and Grammar Aid. University of Notre Dame. Retrieved 2013-03-16{{inconsistent citations}}
- ↑ Wolters Kluwer, Stedman's Medical Dictionary, Wolters Kluwer.
- ↑ Di Bella, Stefano; Ascenzi, Paolo; Siarakas, Steven; Petrosillo, Nicola; di Masi, Alessandra (2016-01-01). "Clostridium difficile Toxins A and B: Insights into Pathogenic Properties and Extraintestinal Effects". Toxins. 8 (5): 134. doi:10.3390/toxins8050134. ISSN 2072-6651. PMC 4885049. PMID 27153087.
- ↑ Just I, Selzer J, von Eichel-Streiber C, Aktories K (1995). "The low molecular mass GTP-binding protein Rh is affected by toxin a from Clostridium difficile". The Journal of Clinical Investigation. 95 (3): 1026–31. doi:10.1172/JCI117747. PMC 441436. PMID 7883950.
- ↑ Barth H, Aktories K, Popoff MR, Stiles BG (2004). "Binary Bacterial Toxins: Biochemistry, Biology, and Applications of Common Clostridium and Bacillus Proteins". Microbiology and Molecular Biology Reviews : MMBR. 68 (3): 373–402, table of contents. doi:10.1128/MMBR.68.3.373-402.2004. PMC 515256. PMID 15353562.
- ↑ [Medical Micriobiology, Fifth Edition, Patrick Murray, Elsevier Mosby, 2005, page 412]
- 1 2 Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, René P, Monczak Y, Dascal A (December 2005). "A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality". The New England Journal of Medicine. 353 (23): 2442–9. doi:10.1056/NEJMoa051639. PMID 16322602.
- ↑ McDonald LC (August 2005). "Clostridium difficile: responding to a new threat from an old enemy" (PDF). Infection Control and Hospital Epidemiology. 26 (8): 672–5. doi:10.1086/502600. PMID 16156321.
- ↑ "Clostridium difficile Infection Information for Patients | HAI | CDC". www.cdc.gov. Retrieved 22 May 2017.
- ↑ "WHO Guidelines on Hand Hygiene in Health Care: a Summary" (PDF). World Health Organization. 2009. p. 31. Retrieved June 18, 2018.
- ↑ Gould, L. Hannah; Limbago, Brandi (2010). "Clostridium difficile in Food and Domestic Animals: A New Foodborne Pathogen?". Clinical Infectious Diseases. 51 (5): 577. doi:10.1086/655692. PMID 20642351.
- ↑ Belluck, Pam (February 25, 2015). "Death Toll From C. Difficile Is Raised". The New York Times. Retrieved 25 February 2015.
- ↑ [Medical Microbiology, Fifth Edition, Patrick Murray, Elsevier Mosby, 2005, page 412]
- ↑ https://www.jmscleaningservices.co.uk/news/7-germs-commonly-found-office-theyre-dangerous/
- 1 2 3 https://www.cdc.gov/hai/organisms/cdiff/cdiff-patient.html
- 1 2 3 Surawicz, Christina M; Brandt, Lawrence J; Binion, David G; Ananthakrishnan, Ashwin N; Curry, Scott R; Gilligan, Peter H; McFarland, Lynne V; Mellow, Mark; Zuckerbraun, Brian S (2013-02-26). "Guidelines for Diagnosis, Treatment and Prevention of Clostridium difficile Infections". The American Journal of Gastroenterology. 108 (4): 478–498. doi:10.1038/ajg.2013.4. ISSN 0002-9270.
- ↑ "Treatment of Recurrent Clostridium difficile Colitis with Vancomycin and Saccharomyces boulardii" (PDF). The American Journal of Gastroenterology.
- ↑ Rupnik M, Wilcox MH, Gerding DN (July 2009). "Clostridium difficile infection: New developments in epidemiology and pathogenesis". Nature Reviews. Microbiology. 7 (7): 526–36. doi:10.1038/nrmicro2164. PMID 19528959.
- ↑ Collins, J.; Robinson, C.; Danhof, H.; Knetsch, C. W.; van Leeuwen, H. C.; Lawley, T. D.; Auchtung, J. M.; Britton, R. A. (2018). "Dietary trehalose enhances virulence of epidemic Clostridium difficile". Nature. doi:10.1038/nature25178. ISSN 0028-0836.
- ↑ He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J (April 2010). "Evolutionary dynamics of Clostridium difficile over short and long time scales" (PDF). Proceedings of the National Academy of Sciences of the United States of America. 107 (16): 7527–32. Bibcode:2010PNAS..107.7527H. doi:10.1073/pnas.0914322107. PMC 2867753. PMID 20368420.
- ↑ Scientists map C. difficile strain – Institute of Public Affairs, Montreal
- ↑ Didelot X, Eyre DW, Cule M, Ip CL, Ansari MA, Griffiths D, Vaughan A, O'Connor L, Golubchik T, Batty EM, Piazza P, Wilson DJ, Bowden R, Donnelly PJ, Dingle KE, Wilcox M, Walker AS, Crook DW, A Peto TE, Harding RM (December 2012). "Microevolutionary analysis of Clostridium difficile genomes to investigate transmission" (PDF). Genome Biology. 13 (12): R118. doi:10.1186/gb-2012-13-12-r118. PMC 4056369. PMID 23259504.
- 1 2 Hargreaves KR, Clokie MR (2014). "Clostridium difficile phages: Still difficult?". Frontiers in Microbiology. 5: 184. doi:10.3389/fmicb.2014.00184. PMC 4009436. PMID 24808893.
References
- Pathogen Safety Data Sheets: Infectious Substances – Clostridium Difficile, Public Health Agency, Canada, 10 September 2014.
- Type strain of Clostridium difficile, BacDive - the Bacterial Diversity Metadatabase.