Human genome
The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome, and the mitochondrial genome.[1] Human genomes include both protein-coding DNA genes and noncoding DNA. Haploid human genomes, which are contained in germ cells (the egg and sperm gamete cells created in the meiosis phase of sexual reproduction before fertilization creates a zygote) consist of three billion DNA base pairs, while diploid genomes (found in somatic cells) have twice the DNA content. While there are significant differences among the genomes of human individuals (on the order of 0.1% due to single-nucleotide variants[2] and 0.6% when considering indels),[3] these are considerably smaller than the differences between humans and their closest living relatives, the bonobos and chimpanzees (~1.1% fixed single-nucleotide variants [4] and 4% when including indels).[5]
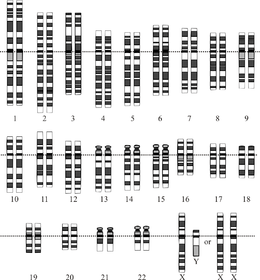 Graphical representation of the idealized human diploid karyotype, showing the organization of the genome into chromosomes. This drawing shows both the female (XX) and male (XY) versions of the 23rd chromosome pair. Chromosomes are shown aligned at their centromeres. The mitochondrial DNA is not shown. | |
| NCBI genome ID | 51 |
|---|---|
| Ploidy | diploid |
| Genome size | 3,234.83 Mbp (mega-basepairs) per haploid genome 6,469.66 Mbp total (diploid). |
| Number of chromosomes | 23 pairs |
The first human genome sequences were published in nearly complete draft form in February 2001 by the Human Genome Project[6] and Celera Corporation.[7] Completion of the Human Genome Project's sequencing effort was announced in 2004 with the publication of a draft genome sequence, leaving just 341 gaps in the sequence, representing highly-repetitive and other DNA that could not be sequenced with the technology available at the time.[8] The human genome was the first of all vertebrates to be sequenced to such near-completion, and as of 2018, the diploid genomes of over a million individual humans had been determined using next-generation sequencing.[9] These data are used worldwide in biomedical science, anthropology, forensics and other branches of science. Such genomic studies have led to advances in the diagnosis and treatment of diseases, and to new insights in many fields of biology, including human evolution.
Although the sequence of the human genome has been (almost) completely determined by DNA sequencing, it is not yet fully understood. Most (though probably not all) genes have been identified by a combination of high throughput experimental and bioinformatics approaches, yet much work still needs to be done to further elucidate the biological functions of their protein and RNA products. Recent results suggest that most of the vast quantities of noncoding DNA within the genome have associated biochemical activities, including regulation of gene expression, organization of chromosome architecture, and signals controlling epigenetic inheritance.
Prior to the acquisition of the full genome sequence, estimates of the number of human genes ranged from 50,000 to 140,000 (with occasional vagueness about whether these estimates included non-protein coding genes).[10] As genome sequence quality and the methods for identifying protein-coding genes improved,[8] the count of recognized protein-coding genes dropped to 19,000-20,000.[11] However, a fuller understanding of the role played by genes expressing regulatory RNAs that do not encode proteins has raised the total number of genes to at least 46,831,[12] plus another 2300 micro-RNA genes.[13] By 2012, functional DNA elements that encode neither RNA nor proteins have been noted.[14] and another 10% equivalent of human genome was found in a recent (2018) population survey.[15] Protein-coding sequences account for only a very small fraction of the genome (approximately 1.5%), and the rest is associated with non-coding RNA genes, regulatory DNA sequences, LINEs, SINEs, introns, and sequences for which as yet no function has been determined.[16]
In June 2016, scientists formally announced HGP-Write, a plan to synthesize the human genome.[17][18]
Molecular organization and gene content
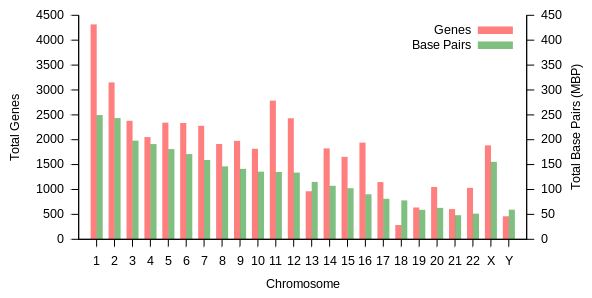
The total length of the human genome is over 3 billion base pairs. The genome is organized into 22 paired chromosomes, termed autosomes, plus the 23rd pair of sex chromosomes (XX) in the female, and (XY) in the male. These are all large linear DNA molecules contained within the cell nucleus. The genome also includes the mitochondrial DNA, a comparatively small circular molecule present in each mitochondrion. Basic information about these molecules and their gene content, based on a reference genome that does not represent the sequence of any specific individual, are provided in the following table. (Data source: Ensembl genome browser release 87, December 2016 for most values; Ensembl genome browser release 68, July 2012 for miRNA, rRNA, snRNA, snoRNA.)
| Chromosome | Length (mm) | Base pairs | Variations | Protein- coding genes | Pseudo- genes | Total long ncRNA | Total small ncRNA | miRNA | rRNA | snRNA | snoRNA | Misc ncRNA | Links | Centromere position (Mbp) | Cumulative (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 85 | 248,956,422 | 12,151,146 | 2058 | 1220 | 1200 | 496 | 134 | 66 | 221 | 145 | 192 | EBI | 125 | 7.9 |
| 2 | 83 | 242,193,529 | 12,945,965 | 1309 | 1023 | 1037 | 375 | 115 | 40 | 161 | 117 | 176 | EBI | 93.3 | 16.2 |
| 3 | 67 | 198,295,559 | 10,638,715 | 1078 | 763 | 711 | 298 | 99 | 29 | 138 | 87 | 134 | EBI | 91 | 23 |
| 4 | 65 | 190,214,555 | 10,165,685 | 752 | 727 | 657 | 228 | 92 | 24 | 120 | 56 | 104 | EBI | 50.4 | 29.6 |
| 5 | 62 | 181,538,259 | 9,519,995 | 876 | 721 | 844 | 235 | 83 | 25 | 106 | 61 | 119 | EBI | 48.4 | 35.8 |
| 6 | 58 | 170,805,979 | 9,130,476 | 1048 | 801 | 639 | 234 | 81 | 26 | 111 | 73 | 105 | EBI | 61 | 41.6 |
| 7 | 54 | 159,345,973 | 8,613,298 | 989 | 885 | 605 | 208 | 90 | 24 | 90 | 76 | 143 | EBI | 59.9 | 47.1 |
| 8 | 50 | 145,138,636 | 8,221,520 | 677 | 613 | 735 | 214 | 80 | 28 | 86 | 52 | 82 | EBI | 45.6 | 52 |
| 9 | 48 | 138,394,717 | 6,590,811 | 786 | 661 | 491 | 190 | 69 | 19 | 66 | 51 | 96 | EBI | 49 | 56.3 |
| 10 | 46 | 133,797,422 | 7,223,944 | 733 | 568 | 579 | 204 | 64 | 32 | 87 | 56 | 89 | EBI | 40.2 | 60.9 |
| 11 | 46 | 135,086,622 | 7,535,370 | 1298 | 821 | 710 | 233 | 63 | 24 | 74 | 76 | 97 | EBI | 53.7 | 65.4 |
| 12 | 45 | 133,275,309 | 7,228,129 | 1034 | 617 | 848 | 227 | 72 | 27 | 106 | 62 | 115 | EBI | 35.8 | 70 |
| 13 | 39 | 114,364,328 | 5,082,574 | 327 | 372 | 397 | 104 | 42 | 16 | 45 | 34 | 75 | EBI | 17.9 | 73.4 |
| 14 | 36 | 107,043,718 | 4,865,950 | 830 | 523 | 533 | 239 | 92 | 10 | 65 | 97 | 79 | EBI | 17.6 | 76.4 |
| 15 | 35 | 101,991,189 | 4,515,076 | 613 | 510 | 639 | 250 | 78 | 13 | 63 | 136 | 93 | EBI | 19 | 79.3 |
| 16 | 31 | 90,338,345 | 5,101,702 | 873 | 465 | 799 | 187 | 52 | 32 | 53 | 58 | 51 | EBI | 36.6 | 82 |
| 17 | 28 | 83,257,441 | 4,614,972 | 1197 | 531 | 834 | 235 | 61 | 15 | 80 | 71 | 99 | EBI | 24 | 84.8 |
| 18 | 27 | 80,373,285 | 4,035,966 | 270 | 247 | 453 | 109 | 32 | 13 | 51 | 36 | 41 | EBI | 17.2 | 87.4 |
| 19 | 20 | 58,617,616 | 3,858,269 | 1472 | 512 | 628 | 179 | 110 | 13 | 29 | 31 | 61 | EBI | 26.5 | 89.3 |
| 20 | 21 | 64,444,167 | 3,439,621 | 544 | 249 | 384 | 131 | 57 | 15 | 46 | 37 | 68 | EBI | 27.5 | 91.4 |
| 21 | 16 | 46,709,983 | 2,049,697 | 234 | 185 | 305 | 71 | 16 | 5 | 21 | 19 | 24 | EBI | 13.2 | 92.6 |
| 22 | 17 | 50,818,468 | 2,135,311 | 488 | 324 | 357 | 78 | 31 | 5 | 23 | 23 | 62 | EBI | 14.7 | 93.8 |
| X | 53 | 156,040,895 | 5,753,881 | 842 | 874 | 271 | 258 | 128 | 22 | 85 | 64 | 100 | EBI | 60.6 | 99.1 |
| Y | 20 | 57,227,415 | 211,643 | 71 | 388 | 71 | 30 | 15 | 7 | 17 | 3 | 8 | EBI | 10.4 | 100 |
| mtDNA | 0.0054 | 16,569 | 929 | 13 | 0 | 0 | 24 | 0 | 2 | 0 | 0 | 0 | EBI | N/A | 100 |
| total | 3,088,286,401 | 155,630,645 | 20412 | 14600 | 14727 | 5037 | 1756 | 532 | 1944 | 1521 | 2213 |
Table 1 (above) summarizes the physical organization and gene content of the human reference genome, with links to the original analysis, as published in the Ensembl database at the European Bioinformatics Institute (EBI) and Wellcome Trust Sanger Institute. Chromosome lengths were estimated by multiplying the number of base pairs by 0.34 nanometers, the distance between base pairs in the DNA double helix. A recent estimation of human chromosome lengths based on updated data reports 205.00 cm for the diploid male genome and 208.23 cm for female, corresponding to weights of 6.41 and 6.51 picograms (pg), respectively.[19] The number of proteins is based on the number of initial precursor mRNA transcripts, and does not include products of alternative pre-mRNA splicing, or modifications to protein structure that occur after translation.
The number of genes in the human genome is not entirely clear because the function of numerous transcripts remains unclear. This is especially true for non-coding RNA (see below). The number of protein-coding genes is better known but there are still on the order of 1,400 questionable genes which may or may not encode functional proteins, usually encoded by short open reading frames. Table 2 gives estimates from various projects and shows these discrepancies.
| Gencode[21] | Ensemble[22] | Refseq[23] | CHESS[24] | |
|---|---|---|---|---|
| protein-coding genes | 19,901 | 20,376 | 20,345 | 21,306 |
| lncRNA genes | 15,779 | 14,720 | 17,712 | 18,484 |
| antisense RNA | 5501 | 28 | 2694 | |
| miscellaneous RNA | 2213 | 2222 | 13,899 | 4347 |
| Pseudogenes | 14,723 | 1740 | 15,952 | |
| total transcripts | 203,835 | 203,903 | 154,484 | 328,827 |
Variations are unique DNA sequence differences that have been identified in the individual human genome sequences analyzed by Ensembl as of December 2016. The number of identified variations is expected to increase as further personal genomes are sequenced and analyzed. In addition to the gene content shown in this table, a large number of non-expressed functional sequences have been identified throughout the human genome (see below). Links open windows to the reference chromosome sequences in the EBI genome browser.
Small non-coding RNAs are RNAs of as many as 200 bases that do not have protein-coding potential. These include: microRNAs, or miRNAs (post-transcriptional regulators of gene expression), small nuclear RNAs, or snRNAs (the RNA components of spliceosomes), and small nucleolar RNAs, or snoRNA (involved in guiding chemical modifications to other RNA molecules). Long non-coding RNAs are RNA molecules longer than 200 bases that do not have protein-coding potential. These include: ribosomal RNAs, or rRNAs (the RNA components of ribosomes), and a variety of other long RNAs that are involved in regulation of gene expression, epigenetic modifications of DNA nucleotides and histone proteins, and regulation of the activity of protein-coding genes. Small discrepancies between total-small-ncRNA numbers and the numbers of specific types of small ncNRAs result from the former values being sourced from Ensembl release 87 and the latter from Ensembl release 68.
Completeness of the human genome sequence
Although the human genome has been completely sequenced for some practical purposes, there are still hundreds of gaps in the sequence and an uncertainty of about 5–10% (300 million basepairs added in 2018).[25] A study, published in 2015, noted more than 160 euchromatic gaps of which 50 gaps were closed.[26] However, there are still numerous gaps in the heterochromatic parts of the genome which is much harder to sequence due to numerous repeats and other intractable sequence features.
Information content
The haploid human genome (23 chromosomes) is about 3 billion base pairs long and contains around 30,000 genes.[27] Since every base pair can be coded by 2 bits, this is about 750 megabytes of data. An individual somatic (diploid) cell contains twice this amount, that is, about 6 billion base pairs. Men have fewer than women because the Y chromosome is about 57 million base pairs whereas the X is about 156 million, but in terms of information men have more because the second X contains almost the same information as the first . Since individual genomes vary in sequence by less than 1% from each other, the variations of a given human's genome from a common reference can be losslessly compressed to roughly 4 megabytes.[28]
The entropy rate of the genome differs significantly between coding and non-coding sequences. It is close to the maximum of 2 bits per base pair for the coding sequences (about 45 million base pairs), but less for the non-coding parts. It ranges between 1.5 and 1.9 bits per base pair for the individual chromosome, except for the Y-chromosome, which has an entropy rate below 0.9 bits per base pair.[29]
Coding vs. noncoding DNA
The content of the human genome is commonly divided into coding and noncoding DNA sequences. Coding DNA is defined as those sequences that can be transcribed into mRNA and translated into proteins during the human life cycle; these sequences occupy only a small fraction of the genome (<2%). Noncoding DNA is made up of all of those sequences (ca. 98% of the genome) that are not used to encode proteins.
Some noncoding DNA contains genes for RNA molecules with important biological functions (noncoding RNA, for example ribosomal RNA and transfer RNA). The exploration of the function and evolutionary origin of noncoding DNA is an important goal of contemporary genome research, including the ENCODE (Encyclopedia of DNA Elements) project, which aims to survey the entire human genome, using a variety of experimental tools whose results are indicative of molecular activity.
Because non-coding DNA greatly outnumbers coding DNA, the concept of the sequenced genome has become a more focused analytical concept than the classical concept of the DNA-coding gene.[30][31]
Coding sequences (protein-coding genes)
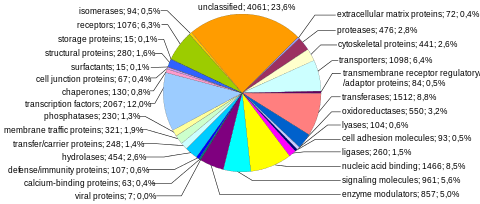
Protein-coding sequences represent the most widely studied and best understood component of the human genome. These sequences ultimately lead to the production of all human proteins, although several biological processes (e.g. DNA rearrangements and alternative pre-mRNA splicing) can lead to the production of many more unique proteins than the number of protein-coding genes. The complete modular protein-coding capacity of the genome is contained within the exome, and consists of DNA sequences encoded by exons that can be translated into proteins. Because of its biological importance, and the fact that it constitutes less than 2% of the genome, sequencing of the exome was the first major milepost of the Human Genome Project.
Number of protein-coding genes. About 20,000 human proteins have been annotated in databases such as Uniprot.[33] Historically, estimates for the number of protein genes have varied widely, ranging up to 2,000,000 in the late 1960s,[34] but several researchers pointed out in the early 1970s that the estimated mutational load from deleterious mutations placed an upper limit of approximately 40,000 for the total number of functional loci (this includes protein-coding and functional non-coding genes).[35] The number of human protein-coding genes is not significantly larger than that of many less complex organisms, such as the roundworm and the fruit fly. This difference may result from the extensive use of alternative pre-mRNA splicing in humans, which provides the ability to build a very large number of modular proteins through the selective incorporation of exons.
Protein-coding capacity per chromosome. Protein-coding genes are distributed unevenly across the chromosomes, ranging from a few dozen to more than 2000, with an especially high gene density within chromosomes 19, 11, and 1 (Table 1). Each chromosome contains various gene-rich and gene-poor regions, which may be correlated with chromosome bands and GC-content.[36] The significance of these nonrandom patterns of gene density is not well understood.[37]
Size of protein-coding genes. The size of protein-coding genes within the human genome shows enormous variability (Table 2). The median size of a protein-coding gene is 26,288 bp (mean = 66,577 bp; Table 2 in[38]). For example, the gene for histone H1a (HIST1HIA) is relatively small and simple, lacking introns and encoding mRNA sequences of 781 nt and a 215 amino acid protein (648 nt open reading frame). Dystrophin (DMD) is the largest protein-coding gene in the human reference genome, spanning a total of 2.2 MB, while Titin (TTN) has the longest coding sequence (114,414 bp), the largest number of exons (363),[39] and the longest single exon (17,106 bp). Over the whole genome, the median size of an exon is 122 bp (mean = 145 bp), the median number of exons is 7 (mean = 8.8), and the median coding sequence encodes 367 amino acids (mean = 447 amino acids; Table 21 in[16]).
| Protein | Chrom | Gene | Length | Exons | Exon length | Intron length | Alt splicing |
|---|---|---|---|---|---|---|---|
| Breast cancer type 2 susceptibility protein | 13 | BRCA2 | 83,736 | 27 | 11,386 | 72,350 | yes |
| Cystic fibrosis transmembrane conductance regulator | 7 | CFTR | 202,881 | 27 | 4,440 | 198,441 | yes |
| Cytochrome b | MT | MTCYB | 1,140 | 1 | 1,140 | 0 | no |
| Dystrophin | X | DMD | 2,220,381 | 79 | 10,500 | 2,209,881 | yes |
| Glyceraldehyde-3-phosphate dehydrogenase | 12 | GAPDH | 4,444 | 9 | 1,425 | 3,019 | yes |
| Hemoglobin beta subunit | 11 | HBB | 1,605 | 3 | 626 | 979 | no |
| Histone H1A | 6 | HIST1H1A | 781 | 1 | 781 | 0 | no |
| Titin | 2 | TTN | 281,434 | 364 | 104,301 | 177,133 | yes |
Table 2. Examples of human protein-coding genes. Chrom, chromosome. Alt splicing, alternative pre-mRNA splicing. (Data source: Ensembl genome browser release 68, July 2012)
Recently, a systematic meta-analysis of updated data of the human genome[38] found that the largest protein-coding gene in the human reference genome is RBFOX1 (RNA binding protein, fox-1 homolog 1), spanning a total of 2.47 MB. Over the whole genome, considering a curated set of protein-coding genes, the median size of an exon is currently estimated to be 133 bp (mean = 309 bp), the median number of exons is currently estimated to be 8 (mean = 11), and the median coding sequence is currently estimated to encode 425 amino acids (mean = 553 amino acids; Tables 2 and 5 in[38]).
Noncoding DNA (ncDNA)
Noncoding DNA is defined as all of the DNA sequences within a genome that are not found within protein-coding exons, and so are never represented within the amino acid sequence of expressed proteins. By this definition, more than 98% of the human genomes is composed of ncDNA.
Numerous classes of noncoding DNA have been identified, including genes for noncoding RNA (e.g. tRNA and rRNA), pseudogenes, introns, untranslated regions of mRNA, regulatory DNA sequences, repetitive DNA sequences, and sequences related to mobile genetic elements.
Numerous sequences that are included within genes are also defined as noncoding DNA. These include genes for noncoding RNA (e.g. tRNA, rRNA), and untranslated components of protein-coding genes (e.g. introns, and 5' and 3' untranslated regions of mRNA).
Protein-coding sequences (specifically, coding exons) constitute less than 1.5% of the human genome.[16] In addition, about 26% of the human genome is introns.[40] Aside from genes (exons and introns) and known regulatory sequences (8–20%), the human genome contains regions of noncoding DNA. The exact amount of noncoding DNA that plays a role in cell physiology has been hotly debated. Recent analysis by the ENCODE project indicates that 80% of the entire human genome is either transcribed, binds to regulatory proteins, or is associated with some other biochemical activity.[14]
It however remains controversial whether all of this biochemical activity contributes to cell physiology, or whether a substantial portion of this is the result transcriptional and biochemical noise, which must be actively filtered out by the organism.[41] Excluding protein-coding sequences, introns, and regulatory regions, much of the non-coding DNA is composed of: Many DNA sequences that do not play a role in gene expression have important biological functions. Comparative genomics studies indicate that about 5% of the genome contains sequences of noncoding DNA that are highly conserved, sometimes on time-scales representing hundreds of millions of years, implying that these noncoding regions are under strong evolutionary pressure and positive selection.[42]
Many of these sequences regulate the structure of chromosomes by limiting the regions of heterochromatin formation and regulating structural features of the chromosomes, such as the telomeres and centromeres. Other noncoding regions serve as origins of DNA replication. Finally several regions are transcribed into functional noncoding RNA that regulate the expression of protein-coding genes (for example[43] ), mRNA translation and stability (see miRNA), chromatin structure (including histone modifications, for example[44] ), DNA methylation (for example[45] ), DNA recombination (for example[46] ), and cross-regulate other noncoding RNAs (for example[47] ). It is also likely that many transcribed noncoding regions do not serve any role and that this transcription is the product of non-specific RNA Polymerase activity.[41]
Pseudogenes
Pseudogenes are inactive copies of protein-coding genes, often generated by gene duplication, that have become nonfunctional through the accumulation of inactivating mutations. Table 1 shows that the number of pseudogenes in the human genome is on the order of 13,000,[48] and in some chromosomes is nearly the same as the number of functional protein-coding genes. Gene duplication is a major mechanism through which new genetic material is generated during molecular evolution.
For example, the olfactory receptor gene family is one of the best-documented examples of pseudogenes in the human genome. More than 60 percent of the genes in this family are non-functional pseudogenes in humans. By comparison, only 20 percent of genes in the mouse olfactory receptor gene family are pseudogenes. Research suggests that this is a species-specific characteristic, as the most closely related primates all have proportionally fewer pseudogenes. This genetic discovery helps to explain the less acute sense of smell in humans relative to other mammals.[49]
Genes for noncoding RNA (ncRNA)
Noncoding RNA molecules play many essential roles in cells, especially in the many reactions of protein synthesis and RNA processing. Noncoding RNA include tRNA, ribosomal RNA, microRNA, snRNA and other non-coding RNA genes including about 60,000 long non-coding RNAs (lncRNAs).[14][50][51][52] Although the number of reported lncRNA genes continues to rise and the exact number in the human genome is yet to be defined, many of them are argued to be non-functional.[53]
Many ncRNAs are critical elements in gene regulation and expression. Noncoding RNA also contributes to epigenetics, transcription, RNA splicing, and the translational machinery. The role of RNA in genetic regulation and disease offers a new potential level of unexplored genomic complexity.[54]
Introns and untranslated regions of mRNA
In addition to the ncRNA molecules that are encoded by discrete genes, the initial transcripts of protein coding genes usually contain extensive noncoding sequences, in the form of introns, 5'-untranslated regions (5'-UTR), and 3'-untranslated regions (3'-UTR). Within most protein-coding genes of the human genome, the length of intron sequences is 10- to 100-times the length of exon sequences (Table 2).
Regulatory DNA sequences
The human genome has many different regulatory sequences which are crucial to controlling gene expression. Conservative estimates indicate that these sequences make up 8% of the genome,[55] however extrapolations from the ENCODE project give that 20[56]-40%[57] of the genome is gene regulatory sequence. Some types of non-coding DNA are genetic "switches" that do not encode proteins, but do regulate when and where genes are expressed (called enhancers).[58]
Regulatory sequences have been known since the late 1960s.[59] The first identification of regulatory sequences in the human genome relied on recombinant DNA technology.[60] Later with the advent of genomic sequencing, the identification of these sequences could be inferred by evolutionary conservation. The evolutionary branch between the primates and mouse, for example, occurred 70–90 million years ago.[61] So computer comparisons of gene sequences that identify conserved non-coding sequences will be an indication of their importance in duties such as gene regulation.[62]
Other genomes have been sequenced with the same intention of aiding conservation-guided methods, for exampled the pufferfish genome.[63] However, regulatory sequences disappear and re-evolve during evolution at a high rate.[64][65][66]
As of 2012, the efforts have shifted toward finding interactions between DNA and regulatory proteins by the technique ChIP-Seq, or gaps where the DNA is not packaged by histones (DNase hypersensitive sites), both of which tell where there are active regulatory sequences in the investigated cell type.[55]
Repetitive DNA sequences
Repetitive DNA sequences comprise approximately 50% of the human genome.[67]
About 8% of the human genome consists of tandem DNA arrays or tandem repeats, low complexity repeat sequences that have multiple adjacent copies (e.g. "CAGCAGCAG...").[68] The tandem sequences may be of variable lengths, from two nucleotides to tens of nucleotides. These sequences are highly variable, even among closely related individuals, and so are used for genealogical DNA testing and forensic DNA analysis.[69]
Repeated sequences of fewer than ten nucleotides (e.g. the dinucleotide repeat (AC)n) are termed microsatellite sequences. Among the microsatellite sequences, trinucleotide repeats are of particular importance, as sometimes occur within coding regions of genes for proteins and may lead to genetic disorders. For example, Huntington's disease results from an expansion of the trinucleotide repeat (CAG)n within the Huntingtin gene on human chromosome 4. Telomeres (the ends of linear chromosomes) end with a microsatellite hexanucleotide repeat of the sequence (TTAGGG)n.
Tandem repeats of longer sequences (arrays of repeated sequences 10–60 nucleotides long) are termed minisatellites.
Mobile genetic elements (transposons) and their relics
Transposable genetic elements, DNA sequences that can replicate and insert copies of themselves at other locations within a host genome, are an abundant component in the human genome. The most abundant transposon lineage, Alu, has about 50,000 active copies,[70] and can be inserted into intragenic and intergenic regions.[71] One other lineage, LINE-1, has about 100 active copies per genome (the number varies between people).[72] Together with non-functional relics of old transposons, they account for over half of total human DNA.[73] Sometimes called "jumping genes", transposons have played a major role in sculpting the human genome. Some of these sequences represent endogenous retroviruses, DNA copies of viral sequences that have become permanently integrated into the genome and are now passed on to succeeding generations.
Mobile elements within the human genome can be classified into LTR retrotransposons (8.3% of total genome), SINEs (13.1% of total genome) including Alu elements, LINEs (20.4% of total genome), SVAs and Class II DNA transposons (2.9% of total genome).
Genomic variation in humans
Human reference genome
With the exception of identical twins, all humans show significant variation in genomic DNA sequences. The human reference genome (HRG) is used as a standard sequence reference.
There are several important points concerning the human reference genome:
- The HRG is a haploid sequence. Each chromosome is represented once.
- The HRG is a composite sequence, and does not correspond to any actual human individual.
- The HRG is periodically updated to correct errors, ambiguities, and unknown "gaps".
- The HRG in no way represents an "ideal" or "perfect" human individual. It is simply a standardized representation or model that is used for comparative purposes.
The Genome Reference Consortium is responsible for updating the HRG. Version 38 was released in December 2013.[74]
Measuring human genetic variation
Most studies of human genetic variation have focused on single-nucleotide polymorphisms (SNPs), which are substitutions in individual bases along a chromosome. Most analyses estimate that SNPs occur 1 in 1000 base pairs, on average, in the euchromatic human genome, although they do not occur at a uniform density. Thus follows the popular statement that "we are all, regardless of race, genetically 99.9% the same",[75] although this would be somewhat qualified by most geneticists. For example, a much larger fraction of the genome is now thought to be involved in copy number variation.[76] A large-scale collaborative effort to catalog SNP variations in the human genome is being undertaken by the International HapMap Project.
The genomic loci and length of certain types of small repetitive sequences are highly variable from person to person, which is the basis of DNA fingerprinting and DNA paternity testing technologies. The heterochromatic portions of the human genome, which total several hundred million base pairs, are also thought to be quite variable within the human population (they are so repetitive and so long that they cannot be accurately sequenced with current technology). These regions contain few genes, and it is unclear whether any significant phenotypic effect results from typical variation in repeats or heterochromatin.
Most gross genomic mutations in gamete germ cells probably result in inviable embryos; however, a number of human diseases are related to large-scale genomic abnormalities. Down syndrome, Turner Syndrome, and a number of other diseases result from nondisjunction of entire chromosomes. Cancer cells frequently have aneuploidy of chromosomes and chromosome arms, although a cause and effect relationship between aneuploidy and cancer has not been established.
Mapping human genomic variation
Whereas a genome sequence lists the order of every DNA base in a genome, a genome map identifies the landmarks. A genome map is less detailed than a genome sequence and aids in navigating around the genome.[77][78]
An example of a variation map is the HapMap being developed by the International HapMap Project. The HapMap is a haplotype map of the human genome, "which will describe the common patterns of human DNA sequence variation."[79] It catalogs the patterns of small-scale variations in the genome that involve single DNA letters, or bases.
Researchers published the first sequence-based map of large-scale structural variation across the human genome in the journal Nature in May 2008.[80][81] Large-scale structural variations are differences in the genome among people that range from a few thousand to a few million DNA bases; some are gains or losses of stretches of genome sequence and others appear as re-arrangements of stretches of sequence. These variations include differences in the number of copies individuals have of a particular gene, deletions, translocations and inversions.
SNP frequency across the human genome
Single-nucleotide polymorphisms (SNPs) do not occur homogeneously across the human genome. In fact, there is enormous diversity in SNP frequency between genes, reflecting different selective pressures on each gene as well as different mutation and recombination rates across the genome. However, studies on SNPs are biased towards coding regions, the data generated from them are unlikely to reflect the overall distribution of SNPs throughout the genome. Therefore, the SNP Consortium protocol was designed to identify SNPs with no bias towards coding regions and the Consortium's 100,000 SNPs generally reflect sequence diversity across the human chromosomes.The SNP Consortium aims to expand the number of SNPs identified across the genome to 300 000 by the end of the first quarter of 2001.[82]
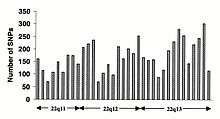
Changes in non-coding sequence and synonymous changes in coding sequence are generally more common than non-synonymous changes, reflecting greater selective pressure reducing diversity at positions dictating amino acid identity. Transitional changes are more common than transversions, with CpG dinucleotides showing the highest mutation rate, presumably due to deamination.
Personal genomes
A personal genome sequence is a (nearly) complete sequence of the chemical base pairs that make up the DNA of a single person. Because medical treatments have different effects on different people due to genetic variations such as single-nucleotide polymorphisms (SNPs), the analysis of personal genomes may lead to personalized medical treatment based on individual genotypes.[83]
The first personal genome sequence to be determined was that of Craig Venter in 2007. Personal genomes had not been sequenced in the public Human Genome Project to protect the identity of volunteers who provided DNA samples. That sequence was derived from the DNA of several volunteers from a diverse population.[84] However, early in the Venter-led Celera Genomics genome sequencing effort the decision was made to switch from sequencing a composite sample to using DNA from a single individual, later revealed to have been Venter himself. Thus the Celera human genome sequence released in 2000 was largely that of one man. Subsequent replacement of the early composite-derived data and determination of the diploid sequence, representing both sets of chromosomes, rather than a haploid sequence originally reported, allowed the release of the first personal genome.[85] In April 2008, that of James Watson was also completed. Since then hundreds of personal genome sequences have been released,[86] including those of Desmond Tutu,[87][88] and of a Paleo-Eskimo.[89] In 2012, the whole genome sequences of two family trios among 1092 genomes was made public.[2] In November 2013, a Spanish family made four personal exome datasets (about 1% of the genome) publicly available under a Creative Commons public domain license.[90][91] The Personal Genome Project (started in 2005) is among the few to make both genome sequences and corresponding medical phenotypes publicly available.[92][93]
The sequencing of individual genomes further unveiled levels of genetic complexity that had not been appreciated before. Personal genomics helped reveal the significant level of diversity in the human genome attributed not only to SNPs but structural variations as well. However, the application of such knowledge to the treatment of disease and in the medical field is only in its very beginnings.[94] Exome sequencing has become increasingly popular as a tool to aid in diagnosis of genetic disease because the exome contributes only 1% of the genomic sequence but accounts for roughly 85% of mutations that contribute significantly to disease.[95]
Human knockouts
In humans, gene knockouts naturally occur as heterozygous or homozygous loss-of-function gene knockouts. These knockouts are often difficult to distinguish, especially within heterogeneous genetic backgrounds. They are also difficult to find as they occur in low frequencies.
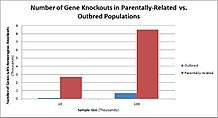
Populations with high rates of consanguinity, such as countries with high rates of first-cousin marriages, display the highest frequencies of homozygous gene knockouts. Such populations include Pakistan, Iceland, and Amish populations. These populations with a high level of parental-relatedness have been subjects of human knock out research which has helped to determine the function of specific genes in humans. By distinguishing specific knockouts, researchers are able to use phenotypic analyses of these individuals to help characterize the gene that has been knocked out.
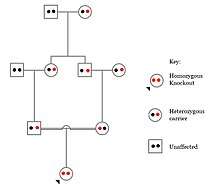
Knockouts in specific genes can cause genetic diseases, potentially have beneficial effects, or even result in no phenotypic effect at all. However, determining a knockout's phenotypic effect and in humans can be challenging. Challenges to characterizing and clinically interpreting knockouts include difficulty calling of DNA variants, determining disruption of protein function (annotation), and considering the amount of influence mosaicism has on the phenotype.[96]
One major study that investigated human knockouts is the Pakistan Risk of Myocardial Infarction study. It was found that individuals possessing a heterozygous loss-of-function gene knockout for the APOC3 gene had lower triglycerides in the blood after consuming a high fat meal as compared to individuals without the mutation. However, individuals possessing homozygous loss-of-function gene knockouts of the APOC3 gene displayed the lowest level of triglycerides in the blood after the fat load test, as they produce no functional APOC3 protein.[97]
Human genetic disorders
Most aspects of human biology involve both genetic (inherited) and non-genetic (environmental) factors. Some inherited variation influences aspects of our biology that are not medical in nature (height, eye color, ability to taste or smell certain compounds, etc.). Moreover, some genetic disorders only cause disease in combination with the appropriate environmental factors (such as diet). With these caveats, genetic disorders may be described as clinically defined diseases caused by genomic DNA sequence variation. In the most straightforward cases, the disorder can be associated with variation in a single gene. For example, cystic fibrosis is caused by mutations in the CFTR gene and is the most common recessive disorder in caucasian populations with over 1,300 different mutations known.[98]
Disease-causing mutations in specific genes are usually severe in terms of gene function and are fortunately rare, thus genetic disorders are similarly individually rare. However, since there are many genes that can vary to cause genetic disorders, in aggregate they constitute a significant component of known medical conditions, especially in pediatric medicine. Molecularly characterized genetic disorders are those for which the underlying causal gene has been identified. Currently there are approximately 2,200 such disorders annotated in the OMIM database.[98]
Studies of genetic disorders are often performed by means of family-based studies. In some instances, population based approaches are employed, particularly in the case of so-called founder populations such as those in Finland, French-Canada, Utah, Sardinia, etc. Diagnosis and treatment of genetic disorders are usually performed by a geneticist-physician trained in clinical/medical genetics. The results of the Human Genome Project are likely to provide increased availability of genetic testing for gene-related disorders, and eventually improved treatment. Parents can be screened for hereditary conditions and counselled on the consequences, the probability of inheritance, and how to avoid or ameliorate it in their offspring.
There are many different kinds of DNA sequence variation, ranging from complete extra or missing chromosomes down to single nucleotide changes. It is generally presumed that much naturally occurring genetic variation in human populations is phenotypically neutral, i.e., has little or no detectable effect on the physiology of the individual (although there may be fractional differences in fitness defined over evolutionary time frames). Genetic disorders can be caused by any or all known types of sequence variation. To molecularly characterize a new genetic disorder, it is necessary to establish a causal link between a particular genomic sequence variant and the clinical disease under investigation. Such studies constitute the realm of human molecular genetics.
With the advent of the Human Genome and International HapMap Project, it has become feasible to explore subtle genetic influences on many common disease conditions such as diabetes, asthma, migraine, schizophrenia, etc. Although some causal links have been made between genomic sequence variants in particular genes and some of these diseases, often with much publicity in the general media, these are usually not considered to be genetic disorders per se as their causes are complex, involving many different genetic and environmental factors. Thus there may be disagreement in particular cases whether a specific medical condition should be termed a genetic disorder.
Additional genetic disorders of mention are Kallman syndrome and Pfeiffer syndrome (gene FGFR1), Fuchs corneal dystrophy (gene TCF4), Hirschsprung's disease (genes RET and FECH), Bardet-Biedl syndrome 1 (genes CCDC28B and BBS1), Bardet-Biedl syndrome 10 (gene BBS10), and facioscapulohumeral muscular dystrophy type 2 (genes D4Z4 and SMCHD1).[99]
Genome sequencing is now able to narrow the genome down to specific locations to more accurately find mutations that will result in a genetic disorder. Copy number variants (CNVs) and single nucleotide variants (SNVs) are also able to be detected at the same time as genome sequencing with newer sequencing procedures available, called Next Generation Sequencing (NGS). This only analyzes a small portion of the genome, around 1-2%. The results of this sequencing can be used for clinical diagnosis of a genetic condition, including Usher syndrome, retinal disease, hearing impairments, diabetes, epilepsy, Leigh disease, hereditary cancers, neuromuscular diseases, primary immunodeficiencies, severe combined immunodeficiency (SCID), and diseases of the mitochondria.[100] NGS can also be used to identify carriers of diseases before conception. The diseases that can be detected in this sequencing include Tay-Sachs disease, Bloom syndrome, Gaucher disease, Canavan disease, familial dysautonomia, cystic fibrosis, spinal muscular atrophy, and fragile-X syndrome. The Next Genome Sequencing can be narrowed down to specifically look for diseases more prevalent in certain ethnic populations.[101]
The categorized table below provides the prevalence as well as the genes or chromosomes associated with some human genetic disorders.
| Disorder | Prevalence | Chromosome or gene involved |
|---|---|---|
| Chromosomal conditions | ||
| Down syndrome | 1:600 | Chromosome 21 |
| Klinefelter syndrome | 1:500–1000 males | Additional X chromosome |
| Turner syndrome | 1:2000 females | Loss of X chromosome |
| Sickle cell anemia | 1 in 50 births in parts of Africa; rarer elsewhere | β-globin (on chromosome 11) |
| Bloom syndrome | 1:48000 Ashkenazi Jews | BLM |
| Cancers | ||
| Breast/Ovarian cancer (susceptibility) | ~5% of cases of these cancer types | BRCA1, BRCA2 |
| FAP (hereditary nonpolyposis coli) | 1:3500 | APC |
| Lynch syndrome | 5–10% of all cases of bowel cancer | MLH1, MSH2, MSH6, PMS2 |
| Fanconi anemia | 1:130000 births | FANCC |
| Neurological conditions | ||
| Huntington disease | 1:20000 | Huntingtin |
| Alzheimer disease ‐ early onset | 1:2500 | PS1, PS2, APP |
| Tay-Sachs | 1:3600 births in Ashkenazi Jews | HEXA gene (on chromosome 15) |
| Canavan disease | 2.5% Eastern European Jewish ancestry | ASPA gene (on chromosome 17) |
| Familial dysautonomia | 600 known cases worldwide since discovery | IKBKAP gene (on chromosome 9) |
| Fragile X syndrome | 1.4:10000 in males, 0.9:10000 in females | FMR1 gene (on X chromosome) |
| Mucolipidosis type IV | 1:90 to 1:100 in Ashkenazi Jews | MCOLN1 |
| Other conditions | ||
| Cystic fibrosis | 1:2500 | CFTR |
| Duchenne muscular dystrophy | 1:3500 boys | Dystrophin |
| Becker muscular dystrophy | 1.5-6:100000 males | DMD |
| Beta thalassemia | 1:100000 | HBB |
| Congenital adrenal hyperplasia | 1:280 in Native Americans and Yupik Eskimos
1:15000 in American Caucasians |
CYP21A2 |
| Glycogen storage disease type I | 1:100000 births in America | G6PC |
| Maple syrup urine disease | 1:180000 in the U.S.
1:176 in Mennonite/Amish communities 1:250000 in Austria |
BCKDHA
BCKDHB DBT DLD |
| Niemann–Pick disease, SMPD1-associated | 1,200 cases worldwide | SMPD1 |
| Usher syndrome | 1:23000 in the U.S.
1:28000 in Norway 1:12500 in Germany |
CDH23
MYO7A PCDH15 USH1C USH1G USH2A GPR98 DFNB31 CLRN1 |
Evolution
Comparative genomics studies of mammalian genomes suggest that approximately 5% of the human genome has been conserved by evolution since the divergence of extant lineages approximately 200 million years ago, containing the vast majority of genes.[102][103] The published chimpanzee genome differs from that of the human genome by 1.23% in direct sequence comparisons.[104] Around 20% of this figure is accounted for by variation within each species, leaving only ~1.06% consistent sequence divergence between humans and chimps at shared genes.[105] This nucleotide by nucleotide difference is dwarfed, however, by the portion of each genome that is not shared, including around 6% of functional genes that are unique to either humans or chimps.[106]
In other words, the considerable observable differences between humans and chimps may be due as much or more to genome level variation in the number, function and expression of genes rather than DNA sequence changes in shared genes. Indeed, even within humans, there has been found to be a previously unappreciated amount of copy number variation (CNV) which can make up as much as 5 – 15% of the human genome. In other words, between humans, there could be +/- 500,000,000 base pairs of DNA, some being active genes, others inactivated, or active at different levels. The full significance of this finding remains to be seen. On average, a typical human protein-coding gene differs from its chimpanzee ortholog by only two amino acid substitutions; nearly one third of human genes have exactly the same protein translation as their chimpanzee orthologs. A major difference between the two genomes is human chromosome 2, which is equivalent to a fusion product of chimpanzee chromosomes 12 and 13.[107] (later renamed to chromosomes 2A and 2B, respectively).
Humans have undergone an extraordinary loss of olfactory receptor genes during our recent evolution, which explains our relatively crude sense of smell compared to most other mammals. Evolutionary evidence suggests that the emergence of color vision in humans and several other primate species has diminished the need for the sense of smell.[108]
In September 2016, scientists reported that, based on human DNA genetic studies, all non-Africans in the world today can be traced to a single population that exited Africa between 50,000 and 80,000 years ago.[109]
Mitochondrial DNA
The human mitochondrial DNA is of tremendous interest to geneticists, since it undoubtedly plays a role in mitochondrial disease. It also sheds light on human evolution; for example, analysis of variation in the human mitochondrial genome has led to the postulation of a recent common ancestor for all humans on the maternal line of descent (see Mitochondrial Eve).
Due to the lack of a system for checking for copying errors, mitochondrial DNA (mtDNA) has a more rapid rate of variation than nuclear DNA. This 20-fold higher mutation rate allows mtDNA to be used for more accurate tracing of maternal ancestry. Studies of mtDNA in populations have allowed ancient migration paths to be traced, such as the migration of Native Americans from Siberia or Polynesians from southeastern Asia . It has also been used to show that there is no trace of Neanderthal DNA in the European gene mixture inherited through purely maternal lineage.[110] Due to the restrictive all or none manner of mtDNA inheritance, this result (no trace of Neanderthal mtDNA) would be likely unless there were a large percentage of Neanderthal ancestry, or there was strong positive selection for that mtDNA (for example, going back 5 generations, only 1 of your 32 ancestors contributed to your mtDNA, so if one of these 32 was pure Neanderthal you would expect that ~3% of your autosomal DNA would be of Neanderthal origin, yet you would have a ~97% chance to have no trace of Neanderthal mtDNA).
Epigenome
Epigenetics describes a variety of features of the human genome that transcend its primary DNA sequence, such as chromatin packaging, histone modifications and DNA methylation, and which are important in regulating gene expression, genome replication and other cellular processes. Epigenetic markers strengthen and weaken transcription of certain genes but do not affect the actual sequence of DNA nucleotides. DNA methylation is a major form of epigenetic control over gene expression and one of the most highly studied topics in epigenetics. During development, the human DNA methylation profile experiences dramatic changes. In early germ line cells, the genome has very low methylation levels. These low levels generally describe active genes. As development progresses, parental imprinting tags lead to increased methylation activity.[111][112]
Epigenetic patterns can be identified between tissues within an individual as well as between individuals themselves. Identical genes that have differences only in their epigenetic state are called epialleles. Epialleles can be placed into three categories: those directly determined by an individual's genotype, those influenced by genotype, and those entirely independent of genotype. The epigenome is also influenced significantly by environmental factors. Diet, toxins, and hormones impact the epigenetic state. Studies in dietary manipulation have demonstrated that methyl-deficient diets are associated with hypomethylation of the epigenome. Such studies establish epigenetics as an important interface between the environment and the genome.[113]
See also
- Genetics
- Genomics
- Genographic Project
- Genomic organization
- Low copy repeats
- Non-coding DNA
- Whole genome sequencing
- Universal Declaration on the Human Genome and Human Rights
References
- Brown TA (2002). "The Human Genome" (2nd ed.). Oxford: Wiley-Liss.
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (November 2012). "An integrated map of genetic variation from 1,092 human genomes". Nature. 491 (7422): 56–65. Bibcode:2012Natur.491...56T. doi:10.1038/nature11632. PMC 3498066. PMID 23128226.
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. (October 2015). "A global reference for human genetic variation". Nature. 526 (7571): 68–74. Bibcode:2015Natur.526...68T. doi:10.1038/nature15393. PMC 4750478. PMID 26432245.
- Chimpanzee Sequencing; Analysis Consortium (2005). "Initial sequence of the chimpanzee genome and comparison with the human genome" (PDF). Nature. 437 (7055): 69–87. Bibcode:2005Natur.437...69.. doi:10.1038/nature04072. PMID 16136131.
- Varki A, Altheide TK (December 2005). "Comparing the human and chimpanzee genomes: searching for needles in a haystack". Genome Research. 15 (12): 1746–58. doi:10.1101/gr.3737405. PMID 16339373.
- International Human Genome Sequencing Consortium Publishes Sequence and Analysis of the Human Genome
- Pennisi E (February 2001). "The human genome". Science. 291 (5507): 1177–80. doi:10.1126/science.291.5507.1177. PMID 11233420.
- International Human Genome Sequencing Consortium (October 2004). "Finishing the euchromatic sequence of the human genome". Nature. 431 (7011): 931–45. Bibcode:2004Natur.431..931H. doi:10.1038/nature03001. PMID 15496913.
- Molteni M (19 November 2018). "Now You Can Sequence Your Whole Genome For Just $200". Wired.
- Wade N (23 September 1999). "Number of Human Genes Is Put at 140,000, a Significant Gain". The New York Times.
- Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, Vazquez J, Valencia A, Tress ML (November 2014). "Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes". Human Molecular Genetics. 23 (22): 5866–78. doi:10.1093/hmg/ddu309. PMC 4204768. PMID 24939910.
- Saey TH (17 September 2018). "A recount of human genes ups the number to at least 46,831". Science News.
- Alles J, Fehlmann T, Fischer U, Backes C, Galata V, Minet M, et al. (April 2019). "An estimate of the total number of true human miRNAs". Nucleic Acids Research. 47 (7): 3353–3364. doi:10.1093/nar/gkz097. PMC 6468295. PMID 30820533.
- Pennisi E (September 2012). "Genomics. ENCODE project writes eulogy for junk DNA". Science. 337 (6099): 1159–1161. doi:10.1126/science.337.6099.1159. PMID 22955811.
- Zhang S (28 November 2018). "300 Million Letters of DNA Are Missing From the Human Genome". The Atlantic.
- International Human Genome Sequencing Consortium (February 2001). "Initial sequencing and analysis of the human genome". Nature. 409 (6822): 860–921. Bibcode:2001Natur.409..860L. doi:10.1038/35057062. PMID 11237011.CS1 maint: uses authors parameter (link)
- Pollack A (2 June 2016). "Scientists Announce HGP-Write, Project to Synthesize the Human Genome". New York Times. Retrieved 2 June 2016.
- Boeke JD, Church G, Hessel A, Kelley NJ, Arkin A, Cai Y, et al. (July 2016). "The Genome Project-Write". Science. 353 (6295): 126–7. Bibcode:2016Sci...353..126B. doi:10.1126/science.aaf6850. PMID 27256881.
- Piovesan A, Pelleri MC, Antonaros F, Strippoli P, Caracausi M, Vitale L (February 2019). "On the length, weight and GC content of the human genome". BMC Research Notes. 12 (1): 106. doi:10.1186/s13104-019-4137-z. PMC 6391780. PMID 30813969.
- Salzberg SL (August 2018). "Open questions: How many genes do we have?". BMC Biology. 16 (1): 94. doi:10.1186/s12915-018-0564-x. PMC 6100717. PMID 30124169.
- "Gencode statistics, version 28". Archived from the original on 2 March 2018. Retrieved 12 July 2018.
- "Ensemble statistics for version 92.38, corresponding to Gencode v28". Retrieved 12 July 2018.
- "NCBI Homo sapiens Annotation Release 108". NIH. 2016.
- "CHESS statistics, version 2.0". Center for Computational Biology. Johns Hopkins University.
- Zhang, Sarah (28 November 2018). "300 Million Letters of DNA Are Missing From the Human Genome". The Atlantic. Retrieved 16 August 2019.
- Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M, Landolin JM, Stamatoyannopoulos JA, Hunkapiller MW, Korlach J, Eichler EE (January 2015). "Resolving the complexity of the human genome using single-molecule sequencing". Nature. 517 (7536): 608–11. Bibcode:2015Natur.517..608C. doi:10.1038/nature13907. PMC 4317254. PMID 25383537.
- "Human Genome Project Completion: Frequently Asked Questions". National Human Genome Research Institute (NHGRI). Retrieved 2 February 2019.
- Christley S, Lu Y, Li C, Xie X (January 2009). "Human genomes as email attachments". Bioinformatics. 25 (2): 274–5. doi:10.1093/bioinformatics/btn582. PMID 18996942.
- Zhandong Liu, Santosh S Venkatesh and Carlo C Maley, Sequence space coverage, entropy of genomes and the potential to detect non-human DNA in human samples, BMC Genomics 2008, 9:509, doi:10.1186/1471-2164-9-509, fig. 6, using the Lempel-Ziv estimators of entropy rate.
- Waters K (7 March 2007). "Molecular Genetics". Stanford Encyclopedia of Philosophy. Retrieved 18 July 2013.
- Gannett L (26 October 2008). "The Human Genome Project". Stanford Encyclopedia of Philosophy. Retrieved 18 July 2013.
- PANTHER Pie Chart at the PANTHER Classification System homepage. Retrieved 25 May 2011
- List of human proteins in the Uniprot Human reference proteome; accessed 28 January 2015
- Kauffman SA (March 1969). "Metabolic stability and epigenesis in randomly constructed genetic nets". Journal of Theoretical Biology. 22 (3): 437–67. doi:10.1016/0022-5193(69)90015-0. PMID 5803332.
- Ohno S (1972). "An argument for the genetic simplicity of man and other mammals". Journal of Human Evolution. 1 (6): 651–662. doi:10.1016/0047-2484(72)90011-5.
- Sémon M, Mouchiroud D, Duret L (February 2005). "Relationship between gene expression and GC-content in mammals: statistical significance and biological relevance". Human Molecular Genetics. 14 (3): 421–7. doi:10.1093/hmg/ddi038. PMID 15590696.
- M. Huang, H. Zhu, B. Shen, G. Gao, "A non-random gait through the human genome", 3rd International Conference on Bioinformatics and Biomedical Engineering (UCBBE, 2009), 1–3
- Piovesan A, Caracausi M, Antonaros F, Pelleri MC, Vitale L (2016). "GeneBase 1.1: a tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics". Database: The Journal of Biological Databases and Curation. 2016: baw153. doi:10.1093/database/baw153. PMC 5199132. PMID 28025344.
- Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S (2001). "The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system". Circulation Research. 89 (11): 1065–72. doi:10.1161/hh2301.100981. PMID 11717165.
- Gregory TR (September 2005). "Synergy between sequence and size in large-scale genomics". Nature Reviews Genetics. 6 (9): 699–708. doi:10.1038/nrg1674. PMID 16151375.
- Palazzo AF, Akef A (June 2012). "Nuclear export as a key arbiter of "mRNA identity" in eukaryotes". Biochimica et Biophysica Acta. 1819 (6): 566–77. doi:10.1016/j.bbagrm.2011.12.012. PMID 22248619.
- Ludwig MZ (December 2002). "Functional evolution of noncoding DNA". Current Opinion in Genetics & Development. 12 (6): 634–9. doi:10.1016/S0959-437X(02)00355-6. PMID 12433575.
- Martens JA, Laprade L, Winston F (June 2004). "Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene". Nature. 429 (6991): 571–4. Bibcode:2004Natur.429..571M. doi:10.1038/nature02538. PMID 15175754.
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY (August 2010). "Long noncoding RNA as modular scaffold of histone modification complexes". Science. 329 (5992): 689–93. Bibcode:2010Sci...329..689T. doi:10.1126/science.1192002. PMC 2967777. PMID 20616235.
- Bartolomei MS, Zemel S, Tilghman SM (May 1991). "Parental imprinting of the mouse H19 gene". Nature. 351 (6322): 153–5. Bibcode:1991Natur.351..153B. doi:10.1038/351153a0. PMID 1709450.
- Kobayashi T, Ganley AR (September 2005). "Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats". Science. 309 (5740): 1581–4. Bibcode:2005Sci...309.1581K. doi:10.1126/science.1116102. PMID 16141077.
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (August 2011). "A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language?". Cell. 146 (3): 353–8. doi:10.1016/j.cell.2011.07.014. PMC 3235919. PMID 21802130.
- Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu XJ, Harte R, Balasubramanian S, Tanzer A, Diekhans M, Reymond A, Hubbard TJ, Harrow J, Gerstein MB (2012). "The GENCODE pseudogene resource". Genome Biology. 13 (9): R51. doi:10.1186/gb-2012-13-9-r51. PMC 3491395. PMID 22951037.
- Gilad Y, Man O, Pääbo S, Lancet D (March 2003). "Human specific loss of olfactory receptor genes". Proceedings of the National Academy of Sciences of the United States of America. 100 (6): 3324–7. Bibcode:2003PNAS..100.3324G. doi:10.1073/pnas.0535697100. PMC 152291. PMID 12612342.
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM (March 2015). "The landscape of long noncoding RNAs in the human transcriptome". Nature Genetics. 47 (3): 199–208. doi:10.1038/ng.3192. PMC 4417758. PMID 25599403.
- Eddy SR (December 2001). "Non-coding RNA genes and the modern RNA world". Nature Reviews Genetics. 2 (12): 919–29. doi:10.1038/35103511. PMID 11733745.
- Managadze D, Lobkovsky AE, Wolf YI, Shabalina SA, Rogozin IB, Koonin EV (2013). "The vast, conserved mammalian lincRNome". PLoS Computational Biology. 9 (2): e1002917. Bibcode:2013PLSCB...9E2917M. doi:10.1371/journal.pcbi.1002917. PMC 3585383. PMID 23468607.
- Palazzo AF, Lee ES (2015). "Non-coding RNA: what is functional and what is junk?". Frontiers in Genetics. 6: 2. doi:10.3389/fgene.2015.00002. PMC 4306305. PMID 25674102.
- Mattick JS, Makunin IV (April 2006). "Non-coding RNA". Human Molecular Genetics. 15 Spec No 1: R17–29. doi:10.1093/hmg/ddl046. PMID 16651366.
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M (September 2012). "An integrated encyclopedia of DNA elements in the human genome". Nature. 489 (7414): 57–74. Bibcode:2012Natur.489...57T. doi:10.1038/nature11247. PMC 3439153. PMID 22955616.
- Birney E (5 September 2012). "ENCODE: My own thoughts". Ewan's Blog: Bioinformatician at large.
- Stamatoyannopoulos JA (September 2012). "What does our genome encode?". Genome Research. 22 (9): 1602–11. doi:10.1101/gr.146506.112. PMC 3431477. PMID 22955972.
- Carroll SB, Gompel N, Prudhomme B (May 2008). "Regulating Evolution". Scientific American. 298 (5): 60–67. Bibcode:2008SciAm.298e..60C. doi:10.1038/scientificamerican0508-60. PMID 18444326.
- Miller JH, Ippen K, Scaife JG, Beckwith JR (1968). "The promoter-operator region of the lac operon of Escherichia coli". J. Mol. Biol. 38 (3): 413–20. doi:10.1016/0022-2836(68)90395-1. PMID 4887877.
- Wright S, Rosenthal A, Flavell R, Grosveld F (1984). "DNA sequences required for regulated expression of beta-globin genes in murine erythroleukemia cells". Cell. 38 (1): 265–73. doi:10.1016/0092-8674(84)90548-8. PMID 6088069.
- Nei M, Xu P, Glazko G (February 2001). "Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms". Proceedings of the National Academy of Sciences of the United States of America. 98 (5): 2497–502. Bibcode:2001PNAS...98.2497N. doi:10.1073/pnas.051611498. PMC 30166. PMID 11226267.
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA (April 2000). "Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons". Science. 288 (5463): 136–40. Bibcode:2000Sci...288..136L. doi:10.1126/science.288.5463.136. PMID 10753117. Summary
- Meunier M. "Genoscope and Whitehead announce a high sequence coverage of the Tetraodon nigroviridis genome". Genoscope. Archived from the original on 16 October 2006. Retrieved 12 September 2006.
- Romero IG, Ruvinsky I, Gilad Y (July 2012). "Comparative studies of gene expression and the evolution of gene regulation". Nature Reviews Genetics. 13 (7): 505–16. doi:10.1038/nrg3229. PMC 4034676. PMID 22705669.
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT (May 2010). "Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding". Science. 328 (5981): 1036–40. Bibcode:2010Sci...328.1036S. doi:10.1126/science.1186176. PMC 3008766. PMID 20378774.
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavaré S, Odom DT (October 2008). "Species-specific transcription in mice carrying human chromosome 21". Science. 322 (5900): 434–8. Bibcode:2008Sci...322..434W. doi:10.1126/science.1160930. PMC 3717767. PMID 18787134.
- Treangen TJ, Salzberg SL (January 2012). "Repetitive DNA and next-generation sequencing: computational challenges and solutions". Nature Reviews Genetics. 13 (1): 36–46. doi:10.1038/nrg3117. PMC 3324860. PMID 22124482.
- Duitama J, Zablotskaya A, Gemayel R, Jansen A, Belet S, Vermeesch JR, Verstrepen KJ, Froyen G (May 2014). "Large-scale analysis of tandem repeat variability in the human genome". Nucleic Acids Research. 42 (9): 5728–41. doi:10.1093/nar/gku212. PMC 4027155. PMID 24682812.
- Pierce BA (2012). Genetics : a conceptual approach (4th ed.). New York: W.H. Freeman. pp. 538–540. ISBN 978-1-4292-3250-0.
- Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE (December 2008). "Active Alu retrotransposons in the human genome". Genome Research. 18 (12): 1875–83. doi:10.1101/gr.081737.108. PMC 2593586. PMID 18836035.
- Liang KH, Yeh CT (2013). "A gene expression restriction network mediated by sense and antisense Alu sequences located on protein-coding messenger RNAs". BMC Genomics. 14: 325. doi:10.1186/1471-2164-14-325. PMC 3655826. PMID 23663499.
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH (April 2003). "Hot L1s account for the bulk of retrotransposition in the human population". Proceedings of the National Academy of Sciences of the United States of America. 100 (9): 5280–5. Bibcode:2003PNAS..100.5280B. doi:10.1073/pnas.0831042100. PMC 154336. PMID 12682288.
- Barton NH, Briggs DE, Eisen JA, Goldstein DB, Patel NH (2007). Evolution. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. ISBN 978-0-87969-684-9.
- NCBI. "GRCh38 – hg38 – Genome – Assembly – NCBI". ncbi.nlm.nih.gov. Retrieved 15 March 2019.
- "from Bill Clinton's 2000 State of the Union address". Archived from the original on 21 February 2017. Retrieved 14 June 2007.
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. (November 2006). "Global variation in copy number in the human genome". Nature. 444 (7118): 444–54. Bibcode:2006Natur.444..444R. doi:10.1038/nature05329. PMC 2669898. PMID 17122850.
- "What's a Genome?". Genomenewsnetwork.org. 15 January 2003. Retrieved 31 May 2009.
- NCBI_user_services (29 March 2004). "Mapping Factsheet". Ncbi.nlm.nih.gov. Archived from the original on 19 July 2010. Retrieved 31 May 2009.
- "About the Project". HapMap. Retrieved 31 May 2009.
- "2008 Release: Researchers Produce First Sequence Map of Large-Scale Structural Variation in the Human Genome". genome.gov. Retrieved 31 May 2009.
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, et al. (May 2008). "Mapping and sequencing of structural variation from eight human genomes". Nature. 453 (7191): 56–64. Bibcode:2008Natur.453...56K. doi:10.1038/nature06862. PMC 2424287. PMID 18451855.
- Gray IC, Campbell DA, Spurr NK (2000). "Single nucleotide polymorphisms as tools in human genetics". Hum. Mol. Genet. 9 (16): 2403–8. doi:10.1093/hmg/9.16.2403. PMID 11005795.
- Lai E (June 2001). "Application of SNP technologies in medicine: lessons learned and future challenges". Genome Research. 11 (6): 927–9. doi:10.1101/gr.192301. PMID 11381021.
- "Human Genome Project Completion: Frequently Asked Questions". genome.gov. Retrieved 31 May 2009.
- Singer E (4 September 2007). "Craig Venter's Genome". MIT Technology Review. Retrieved 25 May 2010.
- "Complete Genomics Adds 29 High-Coverage, Complete Human Genome Sequencing Datasets to Its Public Genomic Repository".
- Sample I (17 February 2010). "Desmond Tutu's genome sequenced as part of genetic diversity study". The Guardian.
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, et al. (2010). "Complete Khoisan and Bantu genomes from southern Africa". Nature. 463 (7283): 943–7. Bibcode:2010Natur.463..943S. doi:10.1038/nature08795. PMC 3890430. PMID 20164927.
- Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, et al. (February 2010). "Ancient human genome sequence of an extinct Palaeo-Eskimo". Nature. 463 (7282): 757–62. Bibcode:2010Natur.463..757R. doi:10.1038/nature08835. PMC 3951495. PMID 20148029.
- Corpas M, Cariaso M, Coletta A, Weiss D, Harrison AP, Moran F, Yang H (12 November 2013). "A Complete Public Domain Family Genomics Dataset". bioRxiv 10.1101/000216.
- Corpas M (June 2013). "Crowdsourcing the corpasome". Source Code for Biology and Medicine. 8 (1): 13. doi:10.1186/1751-0473-8-13. PMC 3706263. PMID 23799911.
- Mao Q, Ciotlos S, Zhang RY, Ball MP, Chin R, Carnevali P, et al. (October 2016). "The whole genome sequences and experimentally phased haplotypes of over 100 personal genomes". GigaScience. 5 (1): 42. doi:10.1186/s13742-016-0148-z. PMC 5057367. PMID 27724973.
- Cai B, Li B, Kiga N, Thusberg J, Bergquist T, Chen YC, et al. (September 2017). "Matching phenotypes to whole genomes: Lessons learned from four iterations of the personal genome project community challenges". Human Mutation. 38 (9): 1266–1276. doi:10.1002/humu.23265. PMC 5645203. PMID 28544481.
- Gonzaga-Jauregui C, Lupski JR, Gibbs RA (2012). "Human genome sequencing in health and disease". Annual Review of Medicine. 63: 35–61. doi:10.1146/annurev-med-051010-162644. PMC 3656720. PMID 22248320.
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP (November 2009). "Genetic diagnosis by whole exome capture and massively parallel DNA sequencing". Proceedings of the National Academy of Sciences of the United States of America. 106 (45): 19096–101. Bibcode:2009PNAS..10619096C. doi:10.1073/pnas.0910672106. PMC 2768590. PMID 19861545.
- Narasimhan VM, Xue Y, Tyler-Smith C (April 2016). "Human Knockout Carriers: Dead, Diseased, Healthy, or Improved?". Trends in Molecular Medicine. 22 (4): 341–351. doi:10.1016/j.molmed.2016.02.006. PMC 4826344. PMID 26988438.
- Saleheen D, Natarajan P, Armean IM, Zhao W, Rasheed A, Khetarpal SA, et al. (April 2017). "Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity". Nature. 544 (7649): 235–239. Bibcode:2017Natur.544..235S. doi:10.1038/nature22034. PMC 5600291. PMID 28406212.
- Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA (January 2002). "Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders". Nucleic Acids Research. 30 (1): 52–5. doi:10.1093/nar/30.1.52. PMID 11752252.
- Katsanis N (November 2016). "The continuum of causality in human genetic disorders". Genome Biology. 17 (1): 233. doi:10.1186/s13059-016-1107-9. PMC 5114767. PMID 27855690.
- Wong, Lee-Jun C. (2017), Wong, Lee-Jun C. (ed.), "Overview of the Clinical Utility of Next Generation Sequencing in Molecular Diagnoses of Human Genetic Disorders", Next Generation Sequencing Based Clinical Molecular Diagnosis of Human Genetic Disorders, Springer International Publishing, pp. 1–11, doi:10.1007/978-3-319-56418-0_1, ISBN 978-3-319-56418-0, retrieved 3 April 2020
- Fedick A, Zhang J (2017). Wong LC (ed.). Next Generation of Carrier Screening. Next Generation Sequencing Based Clinical Molecular Diagnosis of Human Genetic Disorders. Springer International Publishing. pp. 339–354. doi:10.1007/978-3-319-56418-0_16. ISBN 978-3-319-56418-0.
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. (December 2002). "Initial sequencing and comparative analysis of the mouse genome". Nature. 420 (6915): 520–62. Bibcode:2002Natur.420..520W. doi:10.1038/nature01262. PMID 12466850.
the proportion of small (50–100 bp) segments in the mammalian genome that is under (purifying) selection can be estimated to be about 5%. This proportion is much higher than can be explained by protein-coding sequences alone, implying that the genome contains many additional features (such as untranslated regions, regulatory elements, non-protein-coding genes, and chromosomal structural elements) under selection for biological function.
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. (June 2007). "Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project". Nature. 447 (7146): 799–816. Bibcode:2007Natur.447..799B. doi:10.1038/nature05874. PMC 2212820. PMID 17571346.
- The Chimpanzee Sequencing; Analysis Consortium (September 2005). "Initial sequence of the chimpanzee genome and comparison with the human genome". Nature. 437 (7055): 69–87. Bibcode:2005Natur.437...69.. doi:10.1038/nature04072. PMID 16136131.
We calculate the genome-wide nucleotide divergence between human and chimpanzee to be 1.23%, confirming recent results from more limited studies.
- The Chimpanzee Sequencing; Analysis Consortium (September 2005). "Initial sequence of the chimpanzee genome and comparison with the human genome". Nature. 437 (7055): 69–87. Bibcode:2005Natur.437...69.. doi:10.1038/nature04072. PMID 16136131.
we estimate that polymorphism accounts for 14–22% of the observed divergence rate and thus that the fixed divergence is ~1.06% or less
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW (2006). "The evolution of mammalian gene families". PLOS ONE. 1 (1): e85. Bibcode:2006PLoSO...1...85D. doi:10.1371/journal.pone.0000085. PMC 1762380. PMID 17183716.
Our results imply that humans and chimpanzees differ by at least 6% (1,418 of 22,000 genes) in their complement of genes, which stands in stark contrast to the oft-cited 1.5% difference between orthologous nucleotide sequences
- The Chimpanzee Sequencing; Analysis Consortium (September 2005). "Initial sequence of the chimpanzee genome and comparison with the human genome". Nature. 437 (7055): 69–87. Bibcode:2005Natur.437...69.. doi:10.1038/nature04072. PMID 16136131.
Human chromosome 2 resulted from a fusion of two ancestral chromosomes that remained separate in the chimpanzee lineage
Olson MV, Varki A (January 2003). "Sequencing the chimpanzee genome: insights into human evolution and disease". Nature Reviews Genetics. 4 (1): 20–8. doi:10.1038/nrg981. PMID 12509750.Large-scale sequencing of the chimpanzee genome is now imminent.
- Gilad Y, Wiebe V, Przeworski M, Lancet D, Pääbo S (January 2004). "Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates". PLoS Biology. 2 (1): E5. doi:10.1371/journal.pbio.0020005. PMC 314465. PMID 14737185.
Our findings suggest that the deterioration of the olfactory repertoire occurred concomitant with the acquisition of full trichromatic color vision in primates.
- Zimmer C (21 September 2016). "How We Got Here: DNA Points to a Single Migration From Africa". New York Times. Retrieved 22 September 2016.
- Sykes B (9 October 2003). "Mitochondrial DNA and human history". The Human Genome. Archived from the original on 7 September 2015. Retrieved 19 September 2006.
- Misteli T (February 2007). "Beyond the sequence: cellular organization of genome function". Cell. 128 (4): 787–800. doi:10.1016/j.cell.2007.01.028. PMID 17320514.
- Bernstein BE, Meissner A, Lander ES (February 2007). "The mammalian epigenome". Cell. 128 (4): 669–81. doi:10.1016/j.cell.2007.01.033. PMID 17320505.
- Scheen AJ, Junien C (May–June 2012). "[Epigenetics, interface between environment and genes: role in complex diseases]". Revue Médicale de Liège. 67 (5–6): 250–7. PMID 22891475.
External links
| Wikiquote has quotations related to: Human genome |
| Wikimedia Commons has media related to Human chromosomes. |