Spin column-based nucleic acid purification
Spin column-based nucleic acid purification is a solid phase extraction method to quickly purify nucleic acids. This method relies on the fact that nucleic acid will bind to the solid phase of silica under certain conditions.
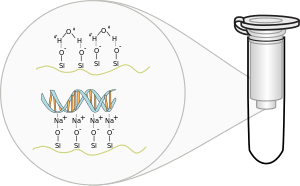
Procedure
The stages of the method are lyse, bind, wash and elute, [1][2] i.e. lysis of cells, binding of nucleic acid to silica gel membrane, washing the nucleic acid bound to the silica gel membrane and elution of the nucleic acid.
To lyse, the cells of a sample are processed with a lytic procedure, which involves breaking the cell membrane and freeing the nucleic acid.
For binding, a buffer solution is then added to the sample along with ethanol or isopropanol. This forms the binding solution. The binding solution is transferred to a spin column and the column is put in a centrifuge. The centrifuge forces the binding solution through a silica gel membrane that is inside the spin column. If the pH and salt concentration of the binding solution are optimal, the nucleic acid will bind to the silica gel membrane as the solution passes through.
To wash, the flow-through is removed and a wash buffer is added to the column. The column is put in a centrifuge again, forcing the wash buffer through the membrane. This removes any remaining impurities from the membrane, leaving only the nucleic acid bound to the silica gel.
To elute, the wash buffer is removed and an elution buffer (or simply water) is added to the column. The column is put in a centrifuge again, forcing the elution buffer through the membrane. The elution buffer removes the nucleic acid from the membrane and the nucleic acid is collected from the bottom of the column.
Related methods
Even prior to the nucleic acid methods employed today it was known that in the presence of chaotropic agents, such as sodium iodide or sodium perchlorate, DNA binds to silica, glass particles or to unicellular algae called diatoms which shield their cell walls with silica. This property was used to purify nucleic acid using glass powder or silica beads under alkaline conditions.[3] This was later improved using guanidinium thiocyanate or guanidinium hydrochloride as the chaotropic agent.[4] The use of glass beads was later changed to silica gel.
See also
- DNA separation by silica adsorption
- Guanidinium thiocyanate-phenol-chloroform extraction
- Ethanol precipitation
- SCODA DNA purification
- Plasmid preparation
References
- Matson, Robert S. (2008). Microarray Methods and Protocols. Boca Raton, Florida: CRC. pp. 27–29. ISBN 978-1420046656.
- Kumar, Anil (2006). Genetic Engineering. New York: Nova Science Publishers. pp. 101–102. ISBN 159454753X.
- Marko MA, Chipperfield R, Birnboim HC. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982 Apr;121(2):382-7. PMID 6179438
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990 Mar;28(3):495-503. PMID 1691208