Translation (biology)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or ER synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is called gene expression.
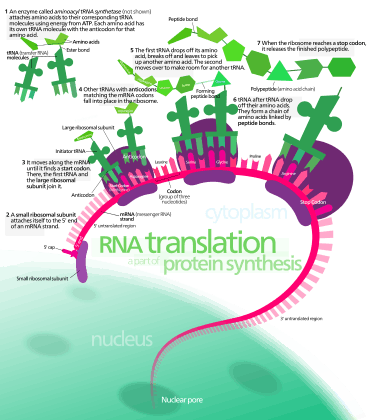
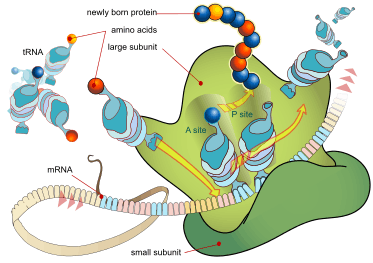
In translation, messenger RNA (mRNA) is decoded in the ribosome decoding center to produce a specific amino acid chain, or polypeptide. The polypeptide later folds into an active protein and performs its functions in the cell. The ribosome facilitates decoding by inducing the binding of complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is read by the ribosome.
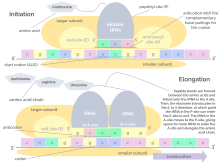
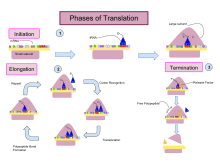
Translation proceeds in three phases:
- Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached at the start codon.
- Elongation: The tRNA transfers an amino acid to the tRNA corresponding to the next codon. The ribosome then moves (translocates) to the next mRNA codon to continue the process, creating an amino acid chain.
- Termination: When a peptidyl tRNA encounters a stop codon, then the ribosome folds the polypeptide into its final structure.
In prokaryotes (unicellular), translation occurs in the cytosol,[1] where the medium and small subunits of the ribosome bind to the tRNA. In eukaryotes, translation occurs in the cytosol or across the membrane of the endoplasmic reticulum in a process called co-translational translocation. In co-translational translocation, the entire ribosome/mRNA complex binds to the outer membrane of the rough endoplasmic reticulum (ER) and the new protein is synthesized and released into the ER; the newly created polypeptide can be stored inside the ER for future vesicle transport and secretion outside the cell, or immediately secreted.
Many types of transcribed RNA, such as transfer RNA, ribosomal RNA, and small nuclear RNA, do not undergo translation into proteins.
A number of antibiotics act by inhibiting translation. These include clindamycin, anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, and puromycin. Prokaryotic ribosomes have a different structure from that of eukaryotic ribosomes, and thus antibiotics can specifically target bacterial infections without any harm to a eukaryotic host's cells.
History
In 1954, Zamecnik and Hoagland discovered tRNA. In 1955, George E. Palade discovered ribosomes.
Basic mechanisms
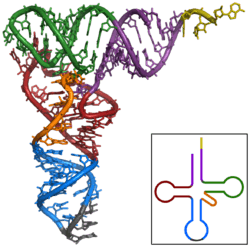
The basic process of translation is the addition of one amino acid at a time to the end of the polypeptide being formed. This process takes place inside the ribosome. A ribosome is made up of a subunit, a small 40S subunit or a large 60S subunit. These subunits come together before translation of mRNA into a protein to provide a location for translation to be carried out and a polypeptide to be produced.[2] The choice of amino acid type to be added is determined by the genetic code on the mRNA molecule. Each amino acid added is matched to a three nucleotide subsequence of the mRNA. For each such triplet possible, the corresponding amino acid is accepted. The successive amino acids added to the chain are matched to successive nucleotide triplets in the mRNA. In this way, the sequence of nucleotides in the template mRNA chain determines the sequence of amino acids in the generated polypeptide.[3] Addition of an amino acid occurs at the C-terminus of the peptide and thus translation is said to be amino-to-carboxyl directed.[4]
The mRNA carries genetic information encoded as a DNA sequence from the chromosomes to the nucleolus. The ribonucleotides are "read" by translational machinery in a sequence of nucleotide triplets called codons. Each of those triplets codes for a specific amino acid.
The ribosome translates this code to a specific sequence of amino acids. The ribosome is a multi-subunit structure containing rRNA and proteins. It is the "factory" where amino acids are assembled into proteins. tRNAs are small noncoding RNA chains (75-90 nucleotides) that transport amino acids to the ribosome. tRNAs have a site for amino acid attachment, and a site called an anticodon. The anticodon is an RNA triplet complementary to the mRNA triplet that codes for their cargo amino acid.
Aminoacyl tRNA synthetases (enzymes) catalyze the bonding between specific tRNAs and the amino acids that their anticodon sequences call for. The product of this reaction is an aminoacyl-tRNA. In prokaryotes, this aminoacyl-tRNA is carried to the ribosome by EF-Tu, where mRNA codons are matched through complementary base pairing to specific tRNA anticodons. Aminoacyl-tRNA synthetases that mispair tRNAs with the wrong amino acids can produce mischarged aminoacyl-tRNAs, which can result in inappropriate amino acids at the respective position in protein. This "mistranslation"[5] of the genetic code naturally occurs at low levels in most organisms, but certain cellular environments cause an increase in permissive mRNA decoding, sometimes to the benefit of the cell.
The ribosome has three sites for tRNA to bind. They are the aminoacyl site (abbreviated A), the peptidyl site (abbreviated P) and the exit site (abbreviated E). With respect to the mRNA, the three sites are oriented 5’ to 3’ E-P-A, because ribosomes move toward the 3' end of mRNA. The A-site binds the incoming tRNA with the complementary codon on the mRNA. The P-site holds the tRNA with the growing polypeptide chain. The E-site holds the tRNA without its amino acid, and the tRNA is then released. When an aminoacyl-tRNA initially binds to its corresponding codon on the mRNA, it is in the A site. Then, a peptide bond forms between the amino acid of the tRNA in the A site and the amino acid of the charged tRNA in the P site. The growing polypeptide chain is transferred to the tRNA in the A site. Translocation occurs, moving the tRNA in the P site, now without an amino acid, to the E site; the tRNA that was in the A site, now charged with the polypeptide chain, is moved to the P site. The tRNA in the E site leaves and another aminoacyl-tRNA enters the A site to repeat the process.[6]
After the new amino acid is added to the chain, and after the mRNA is released out of the nucleus and into the ribosome's core, the energy provided by the hydrolysis of an ATP bound to the translocase EF-G (in prokaryotes) and eEF-2 (in eukaryotes) moves the ribosome down one codon towards the 3' end. The energy required for translation of proteins is significant. For a protein containing n amino acids, the number of high-energy phosphate bonds required to translate it is 4n+1. The rate of translation varies; it is significantly higher in prokaryotic cells (up to 17-21 amino acid residues per second) than in eukaryotic cells (up to 6-9 amino acid residues per second).[7]
Even though the ribosomes are usually considered accurate, processive machines, the translation process is subject to errors that can lead either to the synthesis of erroneous proteins or to the premature abandonment of translation. The rate of error in synthesizing proteins has been estimated to be between 1/105 and 1/103 misincorporated amino acids, depending on the experimental conditions.[8] The rate of premature translation abandonment, instead, has been estimated to be of the order of magnitude of 10−4 events per translated codon.[9] The correct amino acid is covalently bonded to the correct transfer RNA (tRNA) by amino acyl transferases. The amino acid is joined by its carboxyl group to the 3' OH of the tRNA by an ester bond. When the tRNA has an amino acid linked to it, the tRNA is termed "charged". Initiation involves the small subunit of the ribosome binding to the 5' end of mRNA with the help of initiation factors (IF). In prokaryotes, initiation of protein synthesis involves the recognition of a purine-rich initiation sequence on the mRNA called the Shine-Dalgarno sequence. The Shine-Dalgarno sequence binds to a complementary pyrimidine-rich sequence on the 3' end of the 16S rRNA part of the 30S ribosomal subunit. The binding of these complementary sequences ensures that the 30S ribosomal subunit is bound to the mRNA and is aligned such that the initiation codon is placed in the 30S portion of the P-site. Once the mRNA and 30S subunit are properly bound, an initiation factor brings the initiator tRNA-amino acid complex, f-Met-tRNA, to the 30S P site. The initiation phase is completed once a 50S subunit joins the 30 subunit, forming an active 70S ribosome.[10] Termination of the polypeptide occurs when the A site of the ribosome is occupied by a stop codon (UAA, UAG, or UGA) on the mRNA. tRNA usually cannot recognize or bind to stop codons. Instead, the stop codon induces the binding of a release factor protein[11] (RF1 & RF2) that prompts the disassembly of the entire ribosome/mRNA complex by the hydrolysis of the polypeptide chain from the peptidyl transferase center of the ribosome.[12] Drugs or special sequence motifs on the mRNA can change the ribosomal structure so that near-cognate tRNAs are bound to the stop codon instead of the release factors. In such cases of 'translational readthrough', translation continues until the ribosome encounters the next stop codon.[13]
The process of translation is highly regulated in prokaryotic and eukaryotic organisms. Regulation of translation can impact the global rate of protein synthesis which is closely coupled to the metabolic and proliferative state of a cell. In addition, recent work has revealed that genetic differences and their subsequent expression as mRNAs can also impact translation rate in an RNA-specific manner.[14]
Mathematical modeling of translation
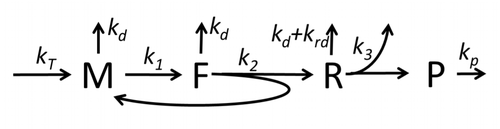
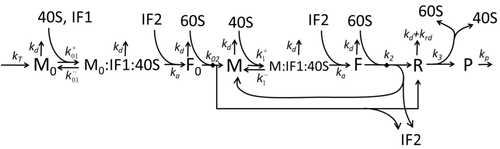
The transcription-translation process description, mentioning only the most basic ”elementary” processes, consists of:
- production of mRNA molecules (including splicing),
- initiation of these molecules with help of initiation factors (e.g., the initiation can include the circularization step though it is not universally required),
- initiation of translation, recruiting the small ribosomal subunit,
- assembly of full ribosomes,
- elongation, i.e. movement of ribosomes along mRNA with production of protein,
- termination of translation,
- degradation of mRNA molecules,
- degradation of proteins.
The process of protein synthesis and translation is a subject of mathematical modeling for a long time starting from the first detailed kinetic models such as [16] or others taking into account stochastic aspects of translation and using computer simulations. Many chemical kinetics-based models of protein synthesis have been developed and analyzed in the last four decades.[17][18] Beyond chemical kinetics, various modeling formalisms such as Totally Asymmetric Simple Exclusion Process (TASEP) [19] Probabilistic Boolean Networks (PBN), Petri Nets and max-plus algebra have been applied to model the detailed kinetics of protein synthesis or some of its stages. A basic model of protein synthesis that took into account all eight 'elementary' processes has been developed,[15] following the paradigm that "useful models are simple and extendable".[20] The simplest model M0 is represented by the reaction kinetic mechanism (Figure M0). It was generalised to include 40S, 60S and initiation factors (IF) binding (Figure M1'). It was extended further to include effect of microRNA on protein synthesis.[21] Most of models in this hierarchy can be solved analytically. These solutions were used to extract 'kinetic signatures' of different specific mechanisms of synthesis regulation.
Genetic code
Whereas other aspects such as the 3D structure, called tertiary structure, of protein can only be predicted using sophisticated algorithms, the amino acid sequence, called primary structure, can be determined solely from the nucleic acid sequence with the aid of a translation table.
This approach may not give the correct amino acid composition of the protein, in particular if unconventional amino acids such as selenocysteine are incorporated into the protein, which is coded for by a conventional stop codon in combination with a downstream hairpin (SElenoCysteine Insertion Sequence, or SECIS).
There are many computer programs capable of translating a DNA/RNA sequence into a protein sequence. Normally this is performed using the Standard Genetic Code, however, few programs can handle all the "special" cases, such as the use of the alternative initiation codons. For instance, the rare alternative start codon CTG codes for methionine when used as a start codon, and for leucine in all other positions.
Example: Condensed translation table for the Standard Genetic Code (from the NCBI Taxonomy webpage).
AAs = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG Starts = ---M---------------M---------------M---------------------------- Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
The "Starts" row indicate three start codons, UUG, CUG, and the very common AUG. It also indicates the first amino acid residue when interpreted as a start: in this case it is all methionine.
Translation tables
Even when working with ordinary eukaryotic sequences such as the yeast genome, it is often desired to be able to use alternative translation tables—namely for translation of the mitochondrial genes. Currently the following translation tables are defined by the NCBI Taxonomy Group for the translation of the sequences in GenBank:[22]
- The standard code
- The vertebrate mitochondrial code
- The yeast mitochondrial code
- The mold, protozoan, and coelenterate mitochondrial code and the mycoplasma/spiroplasma code
- The invertebrate mitochondrial code
- The ciliate, dasycladacean and hexamita nuclear code
- The kinetoplast code
- The echinoderm and flatworm mitochondrial code
- The euplotid nuclear code
- The bacterial, archaeal and plant plastid code
- The alternative yeast nuclear code
- The ascidian mitochondrial code
- The alternative flatworm mitochondrial code
- The Blepharisma nuclear code
- The chlorophycean mitochondrial code
- The trematode mitochondrial code
- The Scenedesmus obliquus mitochondrial code
- The Thraustochytrium mitochondrial code
- The Pterobranchia mitochondrial code
- The candidate division SR1 and gracilibacteria code
- The Pachysolen tannophilus nuclear code
- The karyorelict nuclear code
- The Condylostoma nuclear code
- The Mesodinium nuclear code
- The peritrich nuclear code
- The Blastocrithidia nuclear code
- The Cephalodiscidae mitochondrial code
See also
References
- Gualerzi CO, Pon CL (June 1990). "Initiation of mRNA translation in prokaryotes". Biochemistry. 29 (25): 5881–9. doi:10.1021/bi00477a001. PMID 2200518.
- Biology. McGraw hill education. 2014. p. 249. ISBN 978-981-4581-85-1.
- Neill C (1996). Biology (Fourth ed.). The Benjamin/Cummings Publishing Company. pp. 309–310. ISBN 978-0-8053-1940-8.
- Stryer L (2002). Biochemistry (Fifth ed.). W. H. Freeman and Company. p. 826. ISBN 978-0-7167-4684-3.
- Moghal A, Mohler K, Ibba M (November 2014). "Mistranslation of the genetic code". FEBS Letters. 588 (23): 4305–10. doi:10.1016/j.febslet.2014.08.035. PMC 4254111. PMID 25220850.
- Griffiths A (2008). "9". Introduction to Genetic Analysis (9th ed.). New York: W.H. Freeman and Company. pp. 335–339. ISBN 978-0-7167-6887-6.
- Ross JF, Orlowski M (February 1982). "Growth-rate-dependent adjustment of ribosome function in chemostat-grown cells of the fungus Mucor racemosus". Journal of Bacteriology. 149 (2): 650–3. doi:10.1128/JB.149.2.650-653.1982. PMC 216554. PMID 6799491.
- Wohlgemuth I, Pohl C, Mittelstaet J, Konevega AL, Rodnina MV (October 2011). "Evolutionary optimization of speed and accuracy of decoding on the ribosome". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 366 (1580): 2979–86. doi:10.1098/rstb.2011.0138. PMC 3158919. PMID 21930591.
- Sin C, Chiarugi D, Valleriani A (April 2016). "Quantitative assessment of ribosome drop-off in E. coli". Nucleic Acids Research. 44 (6): 2528–37. doi:10.1093/nar/gkw137. PMC 4824120. PMID 26935582.
- Nakamoto, T (2011). "Mechanisms of the initiation of protein synthesis: In reading frame binding of ribosomes to mRNA". Molecular Biology Reports. 38 (2): 847–855. doi:10.1007/s11033-010-0176-1. PMID 20467902. S2CID 22038744.
- Baggett NE, Zhang Y, Gross CA (March 2017). Ibba M (ed.). "Global analysis of translation termination in E. coli". PLOS Genetics. 13 (3): e1006676. doi:10.1371/journal.pgen.1006676. PMC 5373646. PMID 28301469.
- Mora L, Zavialov A, Ehrenberg M, Buckingham RH (December 2003). "Stop codon recognition and interactions with peptide release factor RF3 of truncated and chimeric RF1 and RF2 from Escherichia coli". Molecular Microbiology. 50 (5): 1467–76. doi:10.1046/j.1365-2958.2003.03799.x. PMID 14651631.
- Schueren F, Thoms S (August 2016). "Functional Translational Readthrough: A Systems Biology Perspective". PLOS Genetics. 12 (8): e1006196. doi:10.1371/JOURNAL.PGEN.1006196. PMC 4973966. PMID 27490485.
- Cenik C, Cenik ES, Byeon GW, Grubert F, Candille SI, Spacek D, Alsallakh B, Tilgner H, Araya CL, Tang H, Ricci E, Snyder MP (November 2015). "Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans". Genome Research. 25 (11): 1610–21. doi:10.1101/gr.193342.115. PMC 4617958. PMID 26297486.
- Gorban, A. N.; Harel-Bellan, A.; Morozova, N.; Zinovyev, A. (2019). "Basic, simple and extendable kinetic model of protein synthesis". Mathematical Biosciences and Engineering. 16 (6): 6602–6622. doi:10.3934/mbe.2019329. PMID 31698578.
- MacDonald, C. T.; Gibbs, J. H.; Pipkin, A. C. (1968). "Kinetics of biopolymerization on nucleic acid templates". Biopolymers. 6 (1): 1–25. doi:10.1002/bip.1968.360060102. PMID 5641411.
- Heinrich, R.; Rapoport, T. A. (1980). "Mathematical modelling of translation of mRNA in eucaryotes; steady states, time-dependent processes and application to reticulocytest". J. Theor. Biol. 86 (2): 279–313. doi:10.1016/0022-5193(80)90008-9. PMID 7442295.
- Skjndal-Bara, N.; Morrisb, D. R. (2007). "Dynamic Model of the Process of Protein Synthesis in Eukaryotic Cells". Bull. Math. Biol. 69 (1): 361–393. doi:10.1007/s11538-006-9128-2. PMID 17031456. S2CID 83701439.
- Andreev, D.; Arnold, M.; Kiniry, S.; et al. (2018). "TASEP modelling provides a parsimonious explanation for the ability of a single uORF to derepress translation during the integrated stress response". eLife. 7: e32563. doi:10.7554/eLife.32563. PMC 6033536. PMID 29932418.
- Coyte, Katharine Z.; Tabuteau, Hervé; Gaffney, Eamonn A.; Foster, Kevin R.; Durham, William M. (2017). "Reply to Baveye and Darnault: Useful models are simple and extendable". Proceedings of the National Academy of Sciences. 114 (14): E2804–E2805. Bibcode:2017PNAS..114E2804C. doi:10.1073/pnas.1702303114. PMC 5389313. PMID 28341710.
- Morozova, N.; Zinovyev, A.; Nonne, N.; Pritchard, L.-L.; Gorban, A. N.; Harel-Bellan, A. (2012). "Kinetic signatures of microRNA modes of action". RNA. 18 (9): 1635–1655. doi:10.1261/rna.032284.112. PMC 3425779. PMID 22850425.
- Elzanowski, Andrzej; Jim Ostell (7 Jan 2019). "The Genetic Codes". National Center for Biotechnology Information. Retrieved 28 March 2019.
Further reading
- Champe PC, Harvey RA, Ferrier DR (2004). Lippincott's Illustrated Reviews: Biochemistry (3rd ed.). Hagerstwon, MD: Lippincott Williams & Wilkins. ISBN 978-0-7817-2265-0.
- Cox M, Nelson DR, Lehninger AL (2005). Lehninger principles of biochemistry (4th ed.). San Francisco...: W.H. Freeman. ISBN 978-0-7167-4339-2.
- Malys N, McCarthy JE (March 2011). "Translation initiation: variations in the mechanism can be anticipated". Cellular and Molecular Life Sciences. 68 (6): 991–1003. doi:10.1007/s00018-010-0588-z. PMID 21076851. S2CID 31720000.
External links
| Wikimedia Commons has media related to Translation (biology). |