Siphoviridae
Siphoviridae is a family of double-stranded DNA viruses in the order Caudovirales. Bacteria and archaea serve as natural hosts. There are currently 783 species in this family, divided among 265 genera and 13 subfamilies.[2][3] The characteristic structural features of this family are a nonenveloped head and noncontractile tail.
| Siphoviridae | |
|---|---|
 | |
| Typical structure of a siphovirus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Duplodnaviria |
| Kingdom: | Heunggongvirae |
| Phylum: | Uroviricota |
| Class: | Caudoviricetes |
| Order: | Caudovirales |
| Family: | Siphoviridae |
| Genera | |
| Synonyms[1] | |
| |
Structure
Viruses in Siphoviridae are non-enveloped, with icosahedral and head-tail geometries[2] (morphotype B1) or a prolate capsid (morphotype B2), and T=7 symmetry. The diameter is around 60 nm.[2] Members of this family are also characterized by their filamentous, cross-banded, noncontractile tails, usually with short terminal and subterminal fibers. Genomes are double stranded and linear, around 50kb in length,[2] containing about 70 genes. The guanine/cytosine content is usually around 52%.
Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by adsorption into the host cell. Replication follows the replicative transposition model. DNA-templated transcription is the method of transcription. Translation takes place by -1 ribosomal frameshifting, and +1 ribosomal frameshifting. The virus exits the host cell by lysis, and holin/endolysin/spanin proteins.[2] Bacteria and archaea serve as the natural host. Transmission routes are passive diffusion.[2]
Taxonomy
The following subfamilies are recognized:[3]
- Arquatrovirinae
- Bclasvirinae
- Chebruvirinae
- Dclasvirinae
- Dolichocephalovirinae
- Guernseyvirinae
- Langleyhallvirinae
- Mccleskeyvirinae
- Mclasvirinae
- Nclasvirinae
- Nymbaxtervirinae
- Pclasvirinae
- Tybeckvirinae
The following genera are unassigned to a subfamily:[3]
- Abbeymikolonvirus
- Abidjanvirus
- Ahduovirus
- Amigovirus
- Anatolevirus
- Andrewvirus
- Andromedavirus
- Appavirus
- Apricotvirus
- Armstrongvirus
- Attisvirus
- Attoomivirus
- Austintatiousvirus
- Bantamvirus
- Barnyardvirus
- Beetrevirus
- Bernalvirus
- Betterkatzvirus
- Bingvirus
- Biseptimavirus
- Bowservirus
- Bridgettevirus
- Britbratvirus
- Bronvirus
- Brussowvirus
- Casadabanvirus
- Cbastvirus
- Cecivirus
- Ceduovirus
- Ceetrepovirus
- Cequinquevirus
- Chenonavirus
- Cheoctovirus
- Chivirus
- Chunghsingvirus
- Cimpunavirus
- Cinunavirus
- Coetzeevirus
- Coralvirus
- Corndogvirus
- Cronusvirus
- Daredevilvirus
- Decurrovirus
- Delepquintavirus
- Demosthenesvirus
- Detrevirus
- Dhillonvirus
- Dismasvirus
- Doucettevirus
- Edenvirus
- Efquatrovirus
- Eiauvirus
- Eisenstarkvirus
- Elerivirus
- Emalynvirus
- Eyrevirus
- Fairfaxidumvirus
- Farahnazvirus
- Feofaniavirus
- Fibralongavirus
- Franklinbayvirus
- Fromanvirus
- Gaiavirus
- Galaxyvirus
- Galunavirus
- Gamtrevirus
- Gesputvirus
- Getseptimavirus
- Ghobesvirus
- Gilesvirus
- Gillianvirus
- Godonkavirus
- Goodmanvirus
- Gordonvirus
- Gordtnkvirus
- Gorganvirus
- Gorjumvirus
- Gustavvirus
- Harrisonvirus
- Hedwigvirus
- Helsingorvirus
- Hendrixvirus
- Hiyaavirus
- Holosalinivirus
- Homburgvirus
- Ikedavirus
- Ilzatvirus
- Incheonvrus
- Inhavirus
- Jacevirus
- Jenstvirus
- Juiceboxvirus
- Kairosalinivirus
- Kelleziovirus
- Kilunavirus
- Klementvirus
- Kojivirus
- Korravirus
- Kostyavirus
- Krampusvirus
- Kryptosalinivirus
- Lacusarxvirus
- Lambdavirus
- Lanavirus
- Laroyevirus
- Lentavirus
- Liebevirus
- Liefievirus
- Lokivirus
- Lomovskayavirus
- Luckybarnesvirus
- Luckytenvirus
- Lughvirus
- Lwoffvirus
- Magadivirus
- Majavirus
- Manhattanvirus
- Mapvirus
- Mardecavirus
- Marvinvirus
- Maxrubnervirus
- Mementomorivirus
- Metamorphoovirus
- Minunavirus
- Moineauvirus
- Montyvirus
- Mudcatvirus
- Murrayvirus
- Nanhaivirus
- Nazgulvirus
- Neferthenavirus
- Nickievirus
- Nipunavirus
- Nonagvirus
- Nonanavirus
- Nyceiraevirus
- Oengusvirus
- Omegavirus
- Oneupvirus
- Orchidvirus
- Oshimavirus
- Pahexavirus
- Pamexvirus
- Papyrusvirus
- Patiencevirus
- Pbi1virus
- Pepyhexavirus
- Phietavirus
- Phifelvirus
- Picardvirus
- Pikminvirus
- Poushouvirus
- Priunavirus
- Psavirus
- Psimunavirus
- Pulverervirus
- Quhwahvirus
- Raleighvirus
- Ravinvirus
- Rerduovirus
- Rigallicvirus
- Rimavirus
- Rogerhendrixvirus
- Ronaldovirus
- Roufvirus
- Rowavirus
- Ruthyvirus
- Samistivirus
- Samunavirus
- Samwavirus
- Sanovirus
- Sansavirus
- Saphexavirus
- Sashavirus
- Sasvirus
- Saundersvirus
- Scapunavirus
- Schnabeltiervirus
- Schubertvirus
- Septimatrevirus
- Seuratvirus
- Seussvirus
- Sextaecvirus
- Sitaravirus
- Skunavirus
- Slashvirus
- Smoothievirus
- Sonalivirus
- Soupsvirus
- Sourvirus
- Spbetavirus
- Squashvirus
- Stanholtvirus
- Steinhofvirus
- Tankvirus
- Terapinvirus
- Tigunavirus
- Timquatrovirus
- Tinduovirus
- Titanvirus
- Tortellinivirus
- Triavirus
- Trigintaduovirus
- Trinavirus
- Trinevirus
- Trippvirus
- Unahavirus
- Vashvirus
- Vegasvirus
- Vendettavirus
- Vhulanivirus
- Vidquintavirus
- Vieuvirus
- Vividuovirus
- Vojvodinavirus
- Wbetavirus
- Weaselvirus
- Wildcatvirus
- Wilnyevirus
- Wizardvirus
- Woesvirus
- Woodruffvirus
- Xiamenvirus
- Xipdecavirus
- Yangvirus
- Yuavirus
- Yvonnevirus
- Zetavirus
Additionally, there is one species that is unassigned to a subfamily and genus:[3] Rhodococcus virus RGL3.
Proposed genera
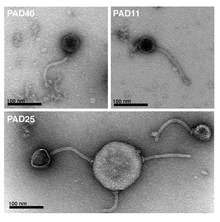
The following genera have been proposed but are not currently ratified by the International Committee on Taxonomy of Viruses:[5][6][7][8][9][10]
- Genus Cba39unalikevirus; type species: Cellulophaga phage phi39:1
- Species:
- Cellulophaga phage phi39:1
- Species:
- Genus Cba46unalikevirus; type species: Cellulophaga phage phi46:1
- Species:
- Cellulophaga phage phi46:1
- Species:
- Genus Cba18unalikevirus; type species: Cellulophaga phage phi18:1
- Species:
- Cellulophaga phage phi18:1
- Cellulophaga phage phi12:1
- Cellulophaga phage phi12:3
- Cellulophaga phage phi17:1
- Cellulophaga phage phi18:2
- Species:
- Genus Cba10unalikevirus; type species: Cellulophaga phage phi10:1
- Species:
- Cellulophaga phage phi10:1
- Cellulophaga phage phi19:1
- Species:
- Genus Cba13unalikevirus; type species: Cellulophaga phage phi13:1
- Species:
- Cellulophaga phage phi13:1
- Cellulophaga phage phi19:2
- Cellulophaga phage phiST
- Species:
- Genus Jk06likevirus; type species: Escherichia phage Jk06
- Species
- Escherichia phage Eb49
- Escherichia phage Jk06
- Escherichia phage Rogue1
- Escherichia phage AHP24
- Escherichia phage AHS24
- Escherichia phage AHP42
- Escherichia phage AKS96
- Enterobacteria phage phiJLA23
- Enterobacteria phage phiKP26
- Species
- Genus Kp36likevirus; type species: Klebsiella phage KP36
- Species
- Enterobacter phage F20
- Klebsiella phage KP36
- Species
- Genus R1tlikevirus; type species: Lactococcus phage r1t
- Species
- Lactococcus phage r1t
- Species
- Genus Rtplikevirus; type species: Escherichia phage Rtp
- Species
- Escherichia phage Rtp
- Enterobacteria phage vB_EcoS_ACG-M12
- Species
- Genus Tlslikevirus; type species: Escherichia phage Tls
- Species
- Escherichia phage Tls
- Salmonella phage FSL SP-126
- Species
Unclassified
In addition to the above viruses, many members of Siphoviridae have been grouped into an unclassified group with no genus assignment.[11] This group includes numerous phages known to infect Lactobacillus, Mycobacterium, Streptococcus, and other bacteria.
References
- Safferman, R.S.; Cannon, R.E.; Desjardins, P.R.; Gromov, B.V.; Haselkorn, R.; Sherman, L.A.; Shilo, M. (1983). "Classification and Nomenclature of Viruses of Cyanobacteria". Intervirology. 19 (2): 61–66. doi:10.1159/000149339. PMID 6408019.
- "Viral Zone". ExPASy. Retrieved 1 July 2015.
- "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 5 May 2020.
- Lood R, Mörgelin M, Holmberg A, Rasmussen M, Collin M (2008). "Inducible Siphoviruses in superficial and deep tissue isolates of Propionibacterium acnes". BMC Microbiol. 8: 139. doi:10.1186/1471-2180-8-139. PMC 2533672. PMID 18702830.
- Proux C, van Sinderen D, Suarez J, Garcia P, Ladero V, Fitzgerald GF, Desiere F, Brüssow H (2002). "The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria". J. Bacteriol. 184 (21): 6026–36. doi:10.1128/JB.184.21.6026-6036.2002. PMC 135392. PMID 12374837.
- Gutiérrez, D; Adriaenssens, EM; Martínez, B; Rodríguez, A; Lavigne, R; Kropinski, AM; García, P (11 September 2013). "Three proposed new bacteriophage genera of staphylococcal phages: "3alikevirus", "77likevirus" and "Phietalikevirus"". Archives of Virology. 159 (2): 389–98. doi:10.1007/s00705-013-1833-1. PMID 24022640.
- Taxonomy Proposals Awaiting Ratification at ICTV
- Taxonomy Proposals Pending at ICTV
- Holmfeldt, K.; Solonenko, N.; Shah, M.; Corrier, K.; Riemann, L.; Verberkmoes, N. C.; Sullivan, M. B. (2013). "Twelve previously unknown phage genera are ubiquitous in global oceans". Proceedings of the National Academy of Sciences. 110 (31): 12798–803. doi:10.1073/pnas.1305956110. PMC 3732932. PMID 23858439.
- Niu, Y. D.; McAllister, T. A.; Nash, J. H. E.; Kropinski, A. M.; Stanford, K. (2014). "Four Escherichia coli O157:H7 Phages: A New Bacteriophage Genus and Taxonomic Classification of T1-Like Phages". PLoS ONE. 9 (6): e100426. doi:10.1371/journal.pone.0100426. PMC 4070988. PMID 24963920.
- "unclassified Siphoviridae". NCBI Taxonomy.
External links
| Wikispecies has information related to Siphoviridae |