Maleate isomerase
In enzymology, a maleate isomerase (EC 5.2.1.1), or maleate cis-tran isomerase, is a member of the Asp/Glu racemase superfamily discovered in bacteria. It is responsible for catalyzing cis-trans isomerization of the C2-C3 double bond in maleate to produce fumarate,[1] which is a critical intermediate in citric acid cycle.[2] The presence of an exogenous mercaptan is required for catalysis to happen.[3]
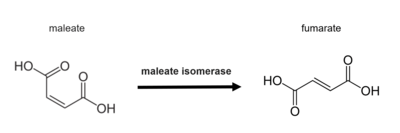 Illustration of the overall isomerization catalyzed by maleate isomerase
Illustration of the overall isomerization catalyzed by maleate isomerase
| Maleate Isomerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Maleate isomerase from Pseudomonas putida | |||||||||
| Identifiers | |||||||||
| EC number | 5.2.1.1 | ||||||||
| CAS number | 9023-74-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Maleate isomerase participates in butanoate metabolism and nicotinate and nicotinamide metabolism.[4] It is an essential enzyme for the last step of metabolic degradation pathway of nicotinic acid. Recently, maleate isomerase has been an industrial target for degradation of tobacco waste.[5][6] It is also got attention for its involvement in aspartic acid and maleic acid production.[7][8][9]
Maleate isomerase has been utilized by multiple bacteria species, including Pseudomonas fluorescens,[3] Alcaligenes faecalis,[10] Bacillus stearothermophilus,[11] Serratia marcescens[8], Pseudomonas putida[12] and Nocardia farcinica.[1][5] The enzyme has a molecular weight of 74,000 and a turnover number of 1,800 moles per mole of protein per min.[3]
Structure
Analogous to other Asp/Glu racemase members, maleate isomerase is formed by two identical protomers, with a flat dimerization surface.[13][14] Each protomer of maleate isomerase has two domains connected by a pseudo-twofold symmetry, with each domain contributes one catalytic cysteine, which is crucial to the isomerase activity at the active site.[5] Experiment shows that substitution of either cysteine by serine significantly reduces the rate of reaction of the enzyme.[1]
In addition to catalytic cysteines, a few other residues at the active site are important for the recognition of the substrate and help stabilize reaction intermediates.[5][1] For example, maleate isomerase from Pseudomonas putida S16 uses Asn17 and Asn169 form hydrogen bonds with the carboxylate group of the maleate distal to Cys82.[5] Tyr139 hydrogen bonds with the carboxylate group of the maleate proximal to Cys82.[5] Pro14 and Val84 make van der Waals interactions with the C2 and C3 carbon atoms of the maleate.[5]
Mechanism
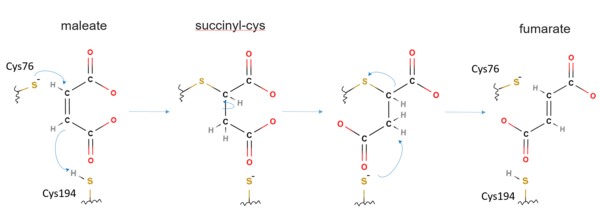
The mechanism of maleate isomerase is considered to be similar to other Asp/Glu racemase members, though have not been fully understood. One proposed reaction mechanism of Nocardia farcinia maleate isomerase is as follows.[1][9] At the active site of maleate isomerase, Cys76 is first deprotonated to be more readily act as a nucleophile.[1] The sulfur atom of the deprotonated Cys76 then carries a direct nucleophilic attack to the C2 atom of the maleate, covalently bonding to the C2 atom.[9][1] Concomitantly, thiol proton of Cys194 is transferred onto the C3 atom of the maleate to form a succinyl-cysteine intermediate.[9][1] The newly formed C2–C3 single bond is then rotated, with Cys76S–C2 bond dissociated, and C3 atom of the maleate deprotonated by Cys194, thus forming fumarate with regeneration of a neutral Cys194.[9][1] In certain type of bacteria, maleate seems completely buried inside the cavity of maleate isomerase and cannot be seen on the surface of the enzyme.[5]
Industrial relevance
Maleate isomerase can be used to produce fumaric acid, an important building block material for polymerization and esterification reactions, from the isomerization of maleic acid.[7] Maleic acid is produced from maleic anhydride.[7]
Maleic acid can also be converted into fumaric acid by thermal or catalytic cis–trans isomerization.[15][16] However, these conversion methods are occurring at high temperatures that causes formation of by-products from maleic and fumaric acids, as a result, yields are below the equilibrium yields.[17] This problem was the main motivation for the alternative enzymatic strategy with maleate isomerase that would facilitate isomerization without by-products.[7]
It is known that, even at moderate temperatures, natural maleate isomerase is unstable.[18] For that reason, heat-stable maleate isomerases are engineered and applied.[7] For example, thermo-stable maleate isomerases derived from Bacillus stearothermophilus, Bacillus brevis, and Bacillus sporothermodurans were used to improve the process.[7][17] In a study using Pseudomonas alcaligenes XD-1, conversion rate from maleic acid into fumaric acid could be achieved as high as 95%.[19][20][7]
References
- Fisch F, Fleites CM, Delenne M, Baudendistel N, Hauer B, Turkenburg JP, Hart S, Bruce NC, Grogan G (August 2010). "A covalent succinylcysteine-like intermediate in the enzyme-catalyzed transformation of maleate to fumarate by maleate isomerase". Journal of the American Chemical Society. 132 (33): 11455–7. doi:10.1021/ja1053576. PMID 20677745.
- Tanaka K, Kobayashi K, Ogasawara N (September 2003). "The Bacillus subtilis YufLM two-component system regulates the expression of the malate transporters MaeN (YufR) and YflS, and is essential for utilization of malate in minimal medium". Microbiology. 149 (Pt 9): 2317–29. doi:10.1099/mic.0.26257-0. PMID 12949159.
- Scher W, Jakoby WB (April 1969). "Maleate isomerase". The Journal of Biological Chemistry. 244 (7): 1878–82. PMID 5780844.
- Behrman EJ, Stanier RY (October 1957). "The bacterial oxidation of nicotinic acid". The Journal of Biological Chemistry. 228 (2): 923–45. PMID 13475371.
- Chen D, Tang H, Lv Y, Zhang Z, Shen K, Lin K, Zhao YL, Wu G, Xu P (March 2013). "Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside". Molecular Microbiology. 87 (6): 1237–44. doi:10.1111/mmi.12163. PMID 23347155.
- Tang H, Yao Y, Wang L, Yu H, Ren Y, Wu G, Xu P (2012). "Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation". Scientific Reports. 2: 377. doi:10.1038/srep00377. PMC 3332521. PMID 22530095.
- Roa Engel CA, Straathof AJ, Zijlmans TW, van Gulik WM, van der Wielen LA (March 2008). "Fumaric acid production by fermentation". Applied Microbiology and Biotechnology. 78 (3): 379–89. doi:10.1007/s00253-007-1341-x. PMC 2243254. PMID 18214471.
- Hatakeyama K, Goto M, Kobayashi M, Terasawa M, Yukawa H (July 2000). "Analysis of oxidation sensitivity of maleate cis-trans isomerase from Serratia marcescens". Bioscience, Biotechnology, and Biochemistry. 64 (7): 1477–85. doi:10.1271/bbb.64.1477. PMID 10945267.
- Dokainish HM, Ion BF, Gauld JW (June 2014). "Computational investigations on the catalytic mechanism of maleate isomerase: the role of the active site cysteine residues". Physical Chemistry Chemical Physics. 16 (24): 12462–74. doi:10.1039/c4cp01342e. PMID 24827730.
- Hatakeyama K, Asai Y, Uchida Y, Kobayashi M, Terasawa M, Yukawa H (October 1997). "Gene cloning and characterization of maleate cis-trans isomerase from Alcaligenes faecalis". Biochemical and Biophysical Research Communications. 239 (1): 74–9. doi:10.1006/bbrc.1997.7430. PMID 9345272.
- Hatakeyama K, Goto M, Uchida Y, Kobayashi M, Terasawa M, Yukawa H (March 2000). "Molecular analysis of maleate cis-trans isomerase from thermophilic bacteria". Bioscience, Biotechnology, and Biochemistry. 64 (3): 569–76. doi:10.1271/bbb.64.569. PMID 10803955.
- Jiménez JI, Canales A, Jiménez-Barbero J, Ginalski K, Rychlewski L, García JL, Díaz E (August 2008). "Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440". Proceedings of the National Academy of Sciences of the United States of America. 105 (32): 11329–34. doi:10.1073/pnas.0802273105. PMC 2516282. PMID 18678916.
- Ruzheinikov SN, Taal MA, Sedelnikova SE, Baker PJ, Rice DW (November 2005). "Substrate-induced conformational changes in Bacillus subtilis glutamate racemase and their implications for drug discovery". Structure. 13 (11): 1707–13. doi:10.1016/j.str.2005.07.024. PMID 16271894.
- Ohtaki A, Nakano Y, Iizuka R, Arakawa T, Yamada K, Odaka M, Yohda M (March 2008). "Structure of aspartate racemase complexed with a dual substrate analogue, citric acid, and implications for the reaction mechanism". Proteins. 70 (4): 1167–74. doi:10.1002/prot.21528. PMID 17847084.
- Lohbeck K, Haferkorn H, Fuhrmann W, Fedtke N (2000). Ullmann's Encyclopedia of Industrial Chemistry. Wiley-VCH Verlag GmbH & Co. KGaA. doi:10.1002/14356007.a16_053. ISBN 978-3-527-30673-2.
- Otsuka K (January 1961). "Cis-trans Isomerase Isomerisation from Maleic Acid to Fumaric Acid". Agricultural and Biological Chemistry. 25 (9): 726–730. doi:10.1271/bbb1961.25.726.
- Goto M, Nara T, Tokumaru I, Fugono N, Uchida Y, Terasawa M (February 1997). "Method of producing fumaric acid". Cite journal requires
|journal=(help) - Takamura Y, Takamura T, Soejima M, Uemura T (January 1969). "Studies on the Induced Synthesis of Maleate Cis-Trans Isomerase by Malonate: Part III. Purification and Properties of Maleate cis-trans Isomerase Induced by Malonate". Journal Agricultural and Biological Chemistry. 33 (5): 718–728. doi:10.1080/00021369.1969.10859369.
- Nakajima-Kambe, Toshiaki; Nozue, Takehiro; Mukouyama, Masaharu; Nakahara, Tadaatsu (January 1997). "Bioconversion of maleic acid to fumaric acid by Pseudomonas alcaligenes strain XD-1". Journal of Fermentation and Bioengineering. 84 (2): 165–168. doi:10.1016/S0922-338X(97)82549-4.
- Ichikawa, Sosaku; Iino, Tomoko; Sato, Seigo; Nakahara, Tadaatsu; Mukataka, Sukekuni (January 2003). "Improvement of production rate and yield of fumaric acid from maleic acid by heat treatment of Pseudomonas alcaligenes strain XD-1". Biochemical Engineering Journal. 13 (1): 7–13. doi:10.1016/S1369-703X(02)00080-3.