CRISPR gene editing
CRISPR gene editing is a genetic engineering technique in molecular biology by which the genomes of living organisms may be modified. It is based on a simplified version of the bacterial CRISPR-Cas9 antiviral defense system. By delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell, the cell's genome can be cut at a desired location, allowing existing genes to be removed and/or new ones added in vivo.[1]
Synopsis
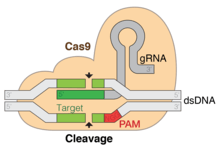
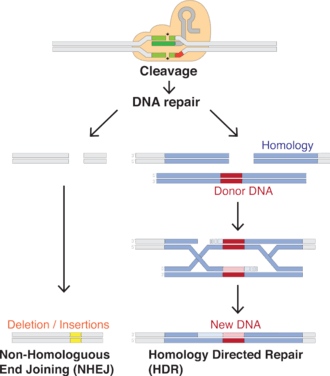
Working like genetic scissors, the Cas9 nuclease opens both strands of the targeted sequence of DNA to introduce the modification by one of two methods. Knock-in mutations, facilitated via homology directed repair (HDR), is the traditional pathway of targeted genomic editing approaches.[2] This allows for the introduction of targeted DNA damage and repair. HDR employs the use of similar DNA sequences to drive the repair of the break via the incorporation of exogenous DNA to function as the repair template.[2] This method relies on the periodic and isolated occurrence of DNA damage at the target site in order for the repair to commence. Knock-out mutations caused by CRISPR-Cas9 result in the repair of the double-stranded break by means of non-homologous end joining (NHEJ). NHEJ can often result in random deletions or insertions at the repair site, which may disrupt or alter gene functionality. Therefore, genomic engineering by CRISPR-Cas9 gives researchers the ability to generate targeted random gene disruption. Because of this, the precision of genome editing is a great concern. Genomic editing leads to irreversible changes to the genome.
While genome editing in eukaryotic cells has been possible using various methods since the 1980s, the methods employed had proved to be inefficient and impractical to implement on a large scale. With the discovery of CRISPR and specifically the Cas9 nuclease molecule, efficient and highly selective editing is now a reality. Cas9 derived from the bacterial species Streptococcus pyogenes has facilitated targeted genomic modification in eukaryotic cells by allowing for a reliable method of creating a targeted break at a specific location as designated by the crRNA and tracrRNA guide strands.[3] The ease with which researchers can insert Cas9 and template RNA in order to silence or cause point mutations at specific loci has proved invaluable to the quick and efficient mapping of genomic models and biological processes associated with various genes in a variety of eukaryotes. Newly engineered variants of the Cas9 nuclease have been developed that significantly reduce off-target activity.[4]
CRISPR-Cas9 genome editing techniques have many potential applications, including in medicine and agriculture. The use of the CRISPR-Cas9-gRNA complex for genome editing[5] was the AAAS's choice for Breakthrough of the Year in 2015.[6] Many bioethical concerns have been raised about the prospect of using CRISPR for germline editing, especially in human embryos.[7]
History
Predecessors
In the early 2000s, researchers developed zinc finger nucleases (ZFNs), synthetic proteins whose DNA-binding domains enable them to create double-stranded breaks in DNA at specific points. In 2010, synthetic nucleases called transcription activator-like effector nucleases (TALENs) provided an easier way to target a double-stranded break to a specific location on the DNA strand. Both zinc finger nucleases and TALENs require the design and creation of a custom protein for each targeted DNA sequence, which is a much more difficult and time-consuming process than that of designing guide RNAs. CRISPRs are much easier to design because the process requires synthesizing only a short RNA sequence, a procedure that is already widely used for many other molecular biology techniques (e.g. creating oligonucleotide primers).[8]
Whereas methods such as RNA interference (RNAi) do not fully suppress gene function, CRISPR, ZFNs, and TALENs provide full irreversible gene knockout.[9] CRISPR can also target several DNA sites simultaneously simply by introducing different gRNAs. In addition, the costs of employing CRISPR are relatively low.[9][10][11]
Patents and commercialization
As of November 2013, SAGE Labs (part of Horizon Discovery group) had exclusive rights from one of those companies to produce and sell genetically engineered rats and non-exclusive rights for mouse and rabbit models.[12] By 2015, Thermo Fisher Scientific had licensed intellectual property from ToolGen to develop CRISPR reagent kits.[13]
As of December 2014, patent rights to CRISPR were contested. Several companies formed to develop related drugs and research tools.[14] As companies ramp up financing, doubts as to whether CRISPR can be quickly monetized were raised.[15] In February 2017 the US Patent Office ruled on a patent interference case brought by University of California with respect to patents issued to the Broad Institute, and found that the Broad patents, with claims covering the application of CRISPR-Cas9 in eukaryotic cells, were distinct from the inventions claimed by University of California.[16][17][18] Shortly after, University of California filed an appeal of this ruling.[19][20]
Recent events
In March 2017, the European Patent Office (EPO) announced its intention to allow broad claims for editing all kinds of cells to Max-Planck Institute in Berlin, University of California, and University of Vienna,[21][22] and in August 2017, the EPO announced its intention to allow CRISPR claims in a patent application that MilliporeSigma had filed.[21] As of August 2017 the patent situation in Europe was complex, with MilliporeSigma, ToolGen, Vilnius University, and Harvard contending for claims, along with University of California and Broad.[23]
In July 2018, the ECJ ruled that gene editing for plants was a sub-category of GMO foods and therefore that the CRISPR technique would henceforth be regulated in the European Union by their rules and regulations for GMOs.[24]
In February 2020, a US trial safely showed CRISPR gene editing on 3 cancer patients.[25]
Genome engineering
CRISPR-Cas9 genome editing is carried out with a Type II CRISPR system. When utilized for genome editing, this system includes Cas9, crRNA, and tracrRNA along with an optional section of DNA repair template that is utilized in either non-homologous end joining (NHEJ) or homology directed repair (HDR).
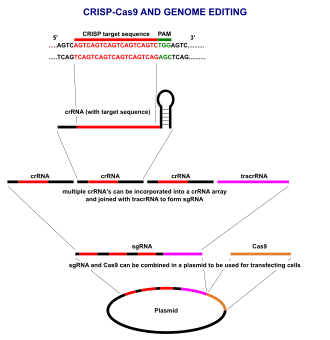
Major components
| Component | Function |
|---|---|
| crRNA | Contains the guide RNA that locates the correct segment of host DNA along with a region that binds to tracrRNA (generally in a hairpin loop form), forming an active complex. |
| tracrRNA | Binds to crRNA and forms an active complex. |
| sgRNA | Single-guide RNAs are a combined RNA consisting of a tracrRNA and at least one crRNA. |
| Cas9 | An enzyme whose active form is able to modify DNA. Many variants exist with different functions (i.e. single-strand nicking, double-strand breaking, DNA binding) due to each enzyme's DNA site recognition function. |
| Repair template | DNA molecule used as a template in the host cell's DNA repair process, allowing insertion of a specific DNA sequence into the host segment broken by Cas9. |
CRISPR-Cas9 often employs a plasmid to transfect the target cells.[26] The main components of this plasmid are displayed in the image and listed in the table. The crRNA is uniquely designed for each application, as this is the sequence that Cas9 uses to identify and directly bind to specific sequences within the host cell's DNA. The crRNA must bind only where editing is desired. The repair template is also uniquely designed for each application, as it must complement to some degree the DNA sequences on either side of the cut and also contain whatever sequence is desired for insertion into the host genome.
Multiple crRNAs and the tracrRNA can be packaged together to form a single-guide RNA (sgRNA).[27] This sgRNA can be included alongside the gene that codes for the Cas9 protein and made into a plasmid in order to be transfected into cells. Many online tools are available to aid in designing effective sgRNA sequences.[28][29]

Structure
CRISPR-Cas9 offers a high degree of fidelity and relatively simple construction. It depends on two factors for its specificity: the target sequence and the protospacer adjacent motif (PAM) sequence. The target sequence is 20 bases long as part of each CRISPR locus in the crRNA array.[26] A typical crRNA array has multiple unique target sequences. Cas9 proteins select the correct location on the host's genome by utilizing the sequence to bond with base pairs on the host DNA. The sequence is not part of the Cas9 protein and as a result is customizable and can be independently synthesized.[30][31]
The PAM sequence on the host genome is recognized by Cas9. Cas9 cannot be easily modified to recognize a different PAM sequence. However, this is ultimately not too limiting, as it is typically a very short and nonspecific sequence that occurs frequently at many places throughout the genome (e.g. the SpCas9 PAM sequence is 5'-NGG-3' and in the human genome occurs roughly every 8 to 12 base pairs).[26]
Once these sequences have been assembled into a plasmid and transfected into cells, the Cas9 protein with the help of the crRNA finds the correct sequence in the host cell's DNA and – depending on the Cas9 variant – creates a single- or double-stranded break at the appropriate location in the DNA.[32]
Properly spaced single-stranded breaks in the host DNA can trigger homology directed repair, which is less error-prone than the non-homologous end joining that typically follows a double-stranded break. Providing a DNA repair template allows for the insertion of a specific DNA sequence at an exact location within the genome. The repair template should extend 40 to 90 base pairs beyond the Cas9-induced DNA break.[26] The goal is for the cell's native HDR process to utilize the provided repair template and thereby incorporate the new sequence into the genome. Once incorporated, this new sequence is now part of the cell's genetic material and passes into its daughter cells.
Delivery
Delivery of Cas9, sgRNA, and associated complexes into cells can occur via viral and non-viral systems. Electroporation of DNA, RNA, or ribonucleocomplexes is a common technique, though it can result in harmful effects on the target cells.[33] Chemical transfection techniques utilizing lipids have also been used to introduce sgRNAs in complex with Cas9 into cells.[34] Types of cells that are more difficult to transfect (e.g. stem cells, neurons, and hematopoietic cells) require more efficient delivery systems, such as those based on lentivirus (LVs), adenovirus (AdV), and adeno-associated virus (AAV).[35][36][37]
Controlled genome editing
Several variants of CRISPR-Cas9 allow gene activation or genome editing with an external trigger such as light or small molecules.[38][39][40] These include photoactivatable CRISPR systems developed by fusing light-responsive protein partners with an activator domain and a dCas9 for gene activation,[41][42] or by fusing similar light-responsive domains with two constructs of split-Cas9,[43][44] or by incorporating caged unnatural amino acids into Cas9,[45] or by modifying the guide RNAs with photocleavable complements for genome editing.[46]
Methods to control genome editing with small molecules include an allosteric Cas9, with no detectable background editing, that will activate binding and cleavage upon the addition of 4-hydroxytamoxifen (4-HT),[38] 4-HT responsive intein-linked Cas9,[47] or a Cas9 that is 4-HT responsive when fused to four ERT2 domains.[48] Intein-inducible split-Cas9 allows dimerization of Cas9 fragments[49] and rapamycin-inducible split-Cas9 system developed by fusing two constructs of split-Cas9 with FRB and FKBP fragments.[50] Other studies have been able to induce transcription of Cas9 with a small molecule, doxycycline.[51][52] Small molecules can also be used to improve homology directed repair,[53] often by inhibiting the non-homologous end joining pathway.[54] These systems allow conditional control of CRISPR activity for improved precision, efficiency, and spatiotemporal control.
CRISPR screening
The clustered regularly interspaced short palindrome repeats (CRISPR)/Cas9 system is a gene-editing technology that can induce double-strand breaks (DSBs), single-strand nicks, or anywhere guide ribonucleic acids (RNAs) can bind with the protospacer adjacent motif (PAM) sequence.[55] By simple changing sequence of gRNA, Cas9-endonuclease can be delivered to a gene of interest and induce DSBs.[56] The efficiency of Cas9-endonuclease and the ease by which genes can be targeted led to the development of CRISPR-knockout (KO) libraries both for mouse and human cells, which can cover either specific gene sets of interest or the whole-genome.[57][58] CRISPR screening helps scientist to create a systematic and high-throughput genetic perturbation within live model organisms. This genetic perturbation is necessary for fully understanding gene function and epigenetic regulation.[59] The advantage of pooled CRISPR libraries is that more genes can be targeted at once.
Knock-out libraries are created in a way to achieve equal representation and performance across all expressed gRNAs and carry an antibiotic or fluorescent selection marker that can be used to recover transduced cells.[55] There are two plasmid systems in CRISPR/Cas9 libraries. First, is all in one plasmid, where sgRNA and Cas9 are produced simultaneously in a transfected cell. Second, is a two-vector system: sgRNA and Cas9 plasmids are delivered separetly.[60] It's important to deliver thousands of unique sgRNAs-containing vectors to a single vessel of cells by viral transduction at low multiplicity of infection (MOI, typically at 0.1-0.6), it prevents the probability that an individual cell clone will get more than one type of sgRNA otherwise it can lead to incorrect assignment of genotype to phenotype.[57]
Once pooled library is prepared it is necessary to carry out a deep sequencing (NGS, next generation sequencing) of PCR-amplifed plasmid DNA in order to reveal abundance of sgRNAs. Cells of interest can be consequentially infected by the library and then selected according to the phenotype. There are 2 types of selection: negative and positive. By negative selection dead or slow growing cells are efficiently detected. It can identify survival-essential genes, which can be further serve as candidates for molecularly targeted drugs. On the other hand, positive selection gives a collection of growth-advantage acquired populations by random mutagenesis.[55] After selection genomic DNA is collected and sequenced by NGS. Depletion or enrichment of sgRNAs is detected and compared to the original sgRNA library, annotated with the target gene that sgRNA corresponds to. Statistical analysis then identify genes that are significantly likely to be relevant to the phenotype of interest.[57]
| Library | ID | Species | PI | Genes targeted | gRNAs per gene | Total gRNAs |
|---|---|---|---|---|---|---|
| Bassik Mouse CRISPR Knockout Library | 1000000121 — 1000000130 | Mouse | Bassik | Varies (∼23,000 in total) | ∼10 | Varies |
| Mouse Tumor Suppressor Gene CRISPR Knockout Library | 113584 EFS backbone
113585 TBG backbone |
Mouse | Chen | 56 | ∼4 | 286 |
| Brie mouse genome-wide library | 73632 (1 plasmid)
73633 (2 plasmid) |
Mouse | Doench and Root | 19,674 | 4 | 78,637 |
| Bassik Human CRISPR Knockout Library | 101926 — 101934 | Human | Bassik | Varies (∼20,500 in total) | ∼10 | Varies |
| Brunello human genome-wide library | 73179 (1 plasmid)
73178 (2 plasmid) |
Human | Doench and Root | 19,114 | 4 | 76,441 |
| Mini-human AsCpf1-based Human Genome-wide Knockout Library | 130630 | Human | Draetta | 16,977 | 3-4 | 17,032 arrays |
Apart from knock-out there are also knock-down (CRISPRi) and activation (CRISPRa) libraries, which using the ability of proteolytically deactivated Cas9-fusion proteins (dCas9) to bind target DNA, which means that gene of interest is not cut but is over-expressed or repressed. It made CRISPR/Cas9 system even more interesting in gene editing. Inactive dCas9 protein modulate gene expression by targeting dCas9-repressors or activators toward promoter or transcriptional start sites of target genes. For repressing genes Cas9 can be fused to KRAB effector domain that makes complex with gRNA, whereas CRISPRa utilizes dCas9 fused to different transcriptional activation domains, which are further directed by gRNA to promoter regions to upregulate expression.[62][63][64]
Applications
Disease models
Cas9 genomic modification has allowed for the quick and efficient generation of transgenic models within the field of genetics. Cas9 can be easily introduced into the target cells along with sgRNA via plasmid transfection in order to model the spread of diseases and the cell's response to and defense against infection.[65] The ability of Cas9 to be introduced in vivo allows for the creation of more accurate models of gene function and mutation effects, all while avoiding the off-target mutations typically observed with older methods of genetic engineering.
The CRISPR and Cas9 revolution in genomic modeling does not extend only to mammals. Traditional genomic models such as Drosophila melanogaster, one of the first model organisms, have seen further refinement in their resolution with the use of Cas9.[65] Cas9 uses cell-specific promoters allowing a controlled use of the Cas9. Cas9 is an accurate method of treating diseases due to the targeting of the Cas9 enzyme only affecting certain cell types. The cells undergoing the Cas9 therapy can also be removed and reintroduced to provide amplified effects of the therapy.[66]
CRISPR-Cas9 can be used to edit the DNA of organisms in vivo and to eliminate individual genes or even entire chromosomes from an organism at any point in its development. Chromosomes that have been successfully deleted in vivo using CRISPR techniques include the Y chromosome and X chromosome of adult lab mice and human chromosomes 14 and 21, in embryonic stem cell lines and aneuploid mice respectively. This method might be useful for treating genetic disorders caused by abnormal numbers of chromosomes, such as Down syndrome and intersex disorders.[67]
Successful in vivo genome editing using CRISPR-Cas9 has been shown in numerous model organisms, including Escherichia coli,[68] Saccharomyces cerevisiae,[69] Candida albicans,[70] Caenorhadbitis elegans,[71] Arabidopsis spp.,[72] Danio rerio,[73] and Mus musculus.[74][75] Successes have been achieved in the study of basic biology, in the creation of disease models,[71] and in the experimental treatment of disease models.[76]
Concerns have been raised that off-target effects (editing of genes besides the ones intended) may confound the results of a CRISPR gene editing experiment (i.e. the observed phenotypic change may not be due to modifying the target gene, but some other gene). Modifications to CRISPR have been made to minimize the possibility of off-target effects. Orthogonal CRISPR experiments are often recommended to confirm the results of a gene editing experiment.[77][78]
CRISPR simplifies the creation of genetically modified organisms for research which mimic disease or show what happens when a gene is knocked down or mutated. CRISPR may be used at the germline level to create organisms in which the targeted gene is changed everywhere (i.e. in all cells/tissues/organs of a multicellular organism), or it may be used in non-germline cells to create local changes that only affect certain cell populations within the organism.[79][80][81]
CRISPR can be utilized to create human cellular models of disease.[82] For instance, when applied to human pluripotent stem cells, CRISPR has been used to introduce targeted mutations in genes relevant to polycystic kidney disease (PKD) and focal segmental glomerulosclerosis (FSGS).[83] These CRISPR-modified pluripotent stem cells were subsequently grown into human kidney organoids that exhibited disease-specific phenotypes. Kidney organoids from stem cells with PKD mutations formed large, translucent cyst structures from kidney tubules. The cysts were capable of reaching macroscopic dimensions, up to one centimeter in diameter.[84] Kidney organoids with mutations in a gene linked to FSGS developed junctional defects between podocytes, the filtering cells affected in that disease. This was traced to the inability of podocytes to form microvilli between adjacent cells.[85] Importantly, these disease phenotypes were absent in control organoids of identical genetic background, but lacking the CRISPR modifications.[83]
A similar approach was taken to model long QT syndrome in cardiomyocytes derived from pluripotent stem cells.[86] These CRISPR-generated cellular models, with isogenic controls, provide a new way to study human disease and test drugs.
Biomedicine
CRISPR-Cas technology has been proposed as a treatment for multiple human diseases, especially those with a genetic cause.[87] Its ability to modify specific DNA sequences makes it a tool with potential to fix disease-causing mutations. Early research in animal models suggest that therapies based on CRISPR technology have potential to treat a wide range of diseases,[88] including cancer,[89] beta-thalassemia,[90] sickle cell disease,[91] hemophilia,[92] cystic fibrosis,[93] Duchenne's muscular dystrophy,[94] Huntington's disease,[95][96] and heart disease.[97] CRISPR may also have applications in tissue engineering and regenerative medicine, such as by creating human blood vessels that lack expression of MHC class II proteins, which often cause transplant rejection.[98]
CRISPR in the treatment of infection
CRISPR-Cas-based "RNA-guided nucleases" can be used to target virulence factors, genes encoding antibiotic resistance, and other medically relevant sequences of interest. This technology thus represents a novel form of antimicrobial therapy and a strategy by which to manipulate bacterial populations.[99][100] Recent studies suggest a correlation between the interfering of the CRISPR-Cas locus and acquisition of antibiotic resistance.[101] This system provides protection of bacteria against invading foreign DNA, such as transposons, bacteriophages, and plasmids. This system was shown to be a strong selective pressure for the acquisition of antibiotic resistance and virulence factor in bacterial pathogens.[101]
Therapies based on CRISPR–Cas3 gene editing technology delivered by engineered bacteriophages could be used to destroy targeted DNA in pathogens.[102] Cas3 is more destructive than the better known Cas9.[103][104]
Research suggests that CRISPR is an effective way to limit replication of multiple herpesviruses. It was able to eradicate viral DNA in the case of Epstein-Barr virus (EBV). Anti-herpesvirus CRISPRs have promising applications such as removing cancer-causing EBV from tumor cells, helping rid donated organs for immunocompromised patients of viral invaders, or preventing cold sore outbreaks and recurrent eye infections by blocking HSV-1 reactivation. As of August 2016, these were awaiting testing.[105]
CRISPR may revive the concept of transplanting animal organs into people. Retroviruses present in animal genomes could harm transplant recipients. In 2015, a team eliminated 62 copies of a particular retroviral DNA sequence from the pig genome in a kidney epithelial cell.[106] Researchers recently demonstrated the ability to birth live pig specimens after removing these retroviruses from their genome using CRISPR for the first time.[107]
CRISPR and cancer
As of 2016 CRISPR had been studied in animal models and cancer cell lines, to learn if it can be used to repair or thwart mutated genes that cause cancer.[108]
The first clinical trial involving CRISPR started in 2016. It involved removing immune cells from people with lung cancer, using CRISPR to edit out the gene expressed PD-1, then administrating the altered cells back to the same person. 20 other trials were under way or nearly ready, mostly in China, as of 2017.[89]
In 2016, the United States Food and Drug Administration (FDA) approved a clinical trial in which CRISPR would be used to alter T cells extracted from people with different kinds of cancer and then administer those engineered T cells back to the same people.[109]
Knockdown/activation

Using "dead" versions of Cas9 (dCas9) eliminates CRISPR's DNA-cutting ability, while preserving its ability to target desirable sequences. Multiple groups added various regulatory factors to dCas9s, enabling them to turn almost any gene on or off or adjust its level of activity.[106] Like RNAi, CRISPR interference (CRISPRi) turns off genes in a reversible fashion by targeting, but not cutting a site. The targeted site is methylated, epigenetically modifying the gene. This modification inhibits transcription. These precisely placed modifications may then be used to regulate the effects on gene expressions and DNA dynamics after the inhibition of certain genome sequences within DNA. Within the past few years, epigenetic marks in different human cells have been closely researched and certain patterns within the marks have been found to correlate with everything ranging from tumor growth to brain activity.[5] Conversely, CRISPR-mediated activation (CRISPRa) promotes gene transcription.[110] Cas9 is an effective way of targeting and silencing specific genes at the DNA level.[111] In bacteria, the presence of Cas9 alone is enough to block transcription. For mammalian applications, a section of protein is added. Its guide RNA targets regulatory DNA sequences called promoters that immediately precede the target gene.[112]
Cas9 was used to carry synthetic transcription factors that activated specific human genes. The technique achieved a strong effect by targeting multiple CRISPR constructs to slightly different locations on the gene's promoter.[112]
RNA editing
In 2016, researchers demonstrated that CRISPR from an ordinary mouth bacterium could be used to edit RNA. The researchers searched databases containing hundreds of millions of genetic sequences for those that resembled CRISPR genes. They considered the fusobacteria Leptotrichia shahii. It had a group of genes that resembled CRISPR genes, but with important differences. When the researchers equipped other bacteria with these genes, which they called C2c2, they found that the organisms gained a novel defense.[113] C2c2 has later been renamed to Cas13a to fit the standard nomenclature for Cas genes.[114]
Many viruses encode their genetic information in RNA rather than DNA that they repurpose to make new viruses. HIV and poliovirus are such viruses. Bacteria with Cas13 make molecules that can dismember RNA, destroying the virus. Tailoring these genes opened any RNA molecule to editing.[113]
CRISPR-Cas systems can also be employed for editing of micro-RNA and long-noncoding RNA genes in plants.[115]
Gene drive
Gene drives may provide a powerful tool to restore balance of ecosystems by eliminating invasive species. Concerns regarding efficacy, unintended consequences in the target species as well as non-target species have been raised particularly in the potential for accidental release from laboratories into the wild. Scientists have proposed several safeguards for ensuring the containment of experimental gene drives including molecular, reproductive, and ecological.[116] Many recommend that immunization and reversal drives be developed in tandem with gene drives in order to overwrite their effects if necessary.[117] There remains consensus that long-term effects must be studied more thoroughly particularly in the potential for ecological disruption that cannot be corrected with reversal drives.[118] As such, DNA computing would be required.
In vitro genetic depletion
Unenriched sequencing libraries often have abundant undesired sequences. Cas9 can specifically deplete the undesired sequences with double strand breakage with up to 99% efficiency and without significant off-target effects as seen with restriction enzymes. Treatment with Cas9 can deplete abundant rRNA while increasing pathogen sensitivity in RNA-seq libraries.[119]
Prime editing
Prime editing[120] (or base editing) is a CRISPR refinement to accurately insert or delete sections of DNA. The CRISPR edits are not always perfect and the cuts can end up in the wrong place. Both issues are a problem for using the technology in medicine.[121] Prime editing does not cut the double-stranded DNA but instead uses the CRISPR targeting apparatus to shuttle an additional enzyme to a desired sequence, where it converts a single nucleotide into another.[122] The new guide, called a pegRNA, contains an RNA template for a new DNA sequence to be added to the genome at the target location. That requires a second protein, attached to Cas9: a reverse transcriptase enzyme, which can make a new DNA strand from the RNA template and insert it at the nicked site.[123] Those three independent pairing events each provide an opportunity to prevent off-target sequences, which significantly increases targeting flexibility and editing precision.[122] Prime editing was developed by researchers at the Broad Institute of MIT and Harvard in Massachusetts.[124] More work is needed to optimize the methods.[124][123]
Society and culture
Human germline modification
As of March 2015, multiple groups had announced ongoing research with the intention of laying the foundations for applying CRISPR to human embryos for human germline engineering, including labs in the US, China, and the UK, as well as US biotechnology company OvaScience.[125] Scientists, including a CRISPR co-discoverer, urged a worldwide moratorium on applying CRISPR to the human germline, especially for clinical use. They said "scientists should avoid even attempting, in lax jurisdictions, germline genome modification for clinical application in humans" until the full implications "are discussed among scientific and governmental organizations".[126][127] These scientists support further low-level research on CRISPR and do not see CRISPR as developed enough for any clinical use in making heritable changes to humans.[128]
In April 2015, Chinese scientists reported results of an attempt to alter the DNA of non-viable human embryos using CRISPR to correct a mutation that causes beta thalassemia, a lethal heritable disorder.[129][130] The study had previously been rejected by both Nature and Science in part because of ethical concerns.[131] The experiments resulted in successfully changing only some of the intended genes, and had off-target effects on other genes. The researchers stated that CRISPR is not ready for clinical application in reproductive medicine.[131] In April 2016, Chinese scientists were reported to have made a second unsuccessful attempt to alter the DNA of non-viable human embryos using CRISPR – this time to alter the CCR5 gene to make the embryo resistant to HIV infection.[132]
In December 2015, an International Summit on Human Gene Editing took place in Washington under the guidance of David Baltimore. Members of national scientific academies of the US, UK, and China discussed the ethics of germline modification. They agreed to support basic and clinical research under certain legal and ethical guidelines. A specific distinction was made between somatic cells, where the effects of edits are limited to a single individual, and germline cells, where genome changes can be inherited by descendants. Heritable modifications could have unintended and far-reaching consequences for human evolution, genetically (e.g. gene-environment interactions) and culturally (e.g. social Darwinism). Altering of gametocytes and embryos to generate heritable changes in humans was defined to be irresponsible. The group agreed to initiate an international forum to address such concerns and harmonize regulations across countries.[133]
In February 2017, the United States National Academies of Sciences, Engineering, and Medicine (NASEM) Committee on Human Gene Editing published a report reviewing ethical, legal, and scientific concerns of genomic engineering technology. The conclusion of the report stated that heritable genome editing is impermissable now but could be justified for certain medical conditions; however, they did not justify the usage of CRISPR for enhancement.[134]
In November 2018, Jiankui He announced that he had edited two human embryos to attempt to disable the gene for CCR5, which codes for a receptor that HIV uses to enter cells. He said that twin girls, Lulu and Nana, had been born a few weeks earlier. He said that the girls still carried functional copies of CCR5 along with disabled CCR5 (mosaicism) and were still vulnerable to HIV. The work was widely condemned as unethical, dangerous, and premature.[135] An international group of scientists called for a global moratorium on genetically editing human embryos.[136]
Policy barriers to genetic engineering
Policy regulations for the CRISPR-Cas9 system vary around the globe. In February 2016, British scientists were given permission by regulators to genetically modify human embryos by using CRISPR-Cas9 and related techniques. However, researchers were forbidden from implanting the embryos and the embryos were to be destroyed after seven days.[137]
The US has an elaborate, interdepartmental regulatory system to evaluate new genetically modified foods and crops. For example, the Agriculture Risk Protection Act of 2000 gives the United States Department of Agriculture the authority to oversee the detection, control, eradication, suppression, prevention, or retardation of the spread of plant pests or noxious weeds to protect the agriculture, environment, and economy of the US. The act regulates any genetically modified organism that utilizes the genome of a predefined "plant pest" or any plant not previously categorized.[138] In 2015, Yinong Yang successfully deactivated 16 specific genes in the white button mushroom to make them non-browning. Since he had not added any foreign-species (transgenic) DNA to his organism, the mushroom could not be regulated by the USDA under Section 340.2.[139] Yang's white button mushroom was the first organism genetically modified with the CRISPR-Cas9 protein system to pass US regulation.[140]
In 2016, the USDA sponsored a committee to consider future regulatory policy for upcoming genetic modification techniques. With the help of the US National Academies of Sciences, Engineering, and Medicine, special interests groups met on April 15 to contemplate the possible advancements in genetic engineering within the next five years and any new regulations that might be needed as a result.[141] In 2017, the Food and Drug Administration proposed a rule that would classify genetic engineering modifications to animals as "animal drugs", subjecting them to strict regulation if offered for sale and reducing the ability for individuals and small businesses to make them profitable.[142][143]
In China, where social conditions sharply contrast with those of the West, genetic diseases carry a heavy stigma.[144] This leaves China with fewer policy barriers to the use of this technology.[145][146]
Recognition
In 2012 and 2013, CRISPR was a runner-up in Science Magazine's Breakthrough of the Year award. In 2015, it was the winner of that award.[106] CRISPR was named as one of MIT Technology Review's 10 breakthrough technologies in 2014 and 2016.[147][148] In 2016, Jennifer Doudna and Emmanuelle Charpentier, along with Rudolph Barrangou, Philippe Horvath, and Feng Zhang won the Gairdner International award. In 2017, Doudna and Charpentier were awarded the Japan Prize in Tokyo, Japan for their revolutionary invention of CRISPR-Cas9. In 2016, Charpentier, Doudna, and Zhang won the Tang Prize in Biopharmaceutical Science.[149]
See also
References
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH (September 2015). "Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells". Nature Biotechnology. 33 (9): 985–9. doi:10.1038/nbt.3290. PMC 4729442. PMID 26121415.
- Bak RO, Gomez-Ospina N, Porteus MH (2018). "Gene Editing on Center Stage". Trends in Genetics. 34 (8): 600–611. doi:10.1016/j.tig.2018.05.004. PMID 29908711.
- Zhang JH, Pandey M, Kahler JF, Loshakov A, Harris B, Dagur PK, Mo YY, Simonds WF (November 2014). "Improving the specificity and efficacy of CRISPR/CAS9 and gRNA through target specific DNA reporter". Journal of Biotechnology. 189: 1–8. doi:10.1016/j.jbiotec.2014.08.033. PMC 4252756. PMID 25193712.
- Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, Bode NM, McNeill MS, Yan S, Camarena J, Lee CM, Park SH, Wiebking V, Bak RO, Gomez-Ospina N, Pavel-Dinu M, Sun W, Bao G, Porteus MH, Behlke MA (August 2018). "A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells". Nature Medicine. 24 (8): 1216–1224. doi:10.1038/s41591-018-0137-0. PMC 6107069. PMID 30082871.
- Ledford H (March 2016). "CRISPR: gene editing is just the beginning". Nature. 531 (7593): 156–9. Bibcode:2016Natur.531..156L. doi:10.1038/531156a. PMID 26961639.
- Travis J (17 December 2015). "Breakthrough of the Year: CRISPR makes the cut". Science Magazine. American Association for the Advancement of Science.
- Ledford H (June 2015). "CRISPR, the disruptor". Nature. 522 (7554): 20–4. Bibcode:2015Natur.522...20L. doi:10.1038/522020a. PMID 26040877.
- Young S (11 February 2014). "CRISPR and Other Genome Editing Tools Boost Medical Research and Gene Therapy's Reach". MIT Technology Review. Retrieved 2014-04-13.
- Heidenreich M, Zhang F (January 2016). "Applications of CRISPR-Cas systems in neuroscience". Nature Reviews. Neuroscience. 17 (1): 36–44. doi:10.1038/nrn.2015.2. PMC 4899966. PMID 26656253.
- Barrangou R, Doudna JA (September 2016). "Applications of CRISPR technologies in research and beyond". Nature Biotechnology. 34 (9): 933–941. doi:10.1038/nbt.3659. PMID 27606440.
- Cox DB, Platt RJ, Zhang F (February 2015). "Therapeutic genome editing: prospects and challenges". Nature Medicine. 21 (2): 121–31. doi:10.1038/nm.3793. PMC 4492683. PMID 25654603.
- "CRISPR Madness". GEN. 2013-11-08.
- Staff (1 April 2015). "News: Products & Services". Genetic Engineering & Biotechnology News (Paper). 35 (7): 8. doi:10.1089/gen.35.21.05.
- "Who Owns the Biggest Biotech Discovery of the Century? There's a bitter fight over the patents for CRISPR, a breakthrough new form of DNA editing". MIT Technology Review. Retrieved 25 February 2015.
- Fye S. "Genetic Rough Draft: Editas and CRISPR". The Atlas Business Journal. Retrieved 19 January 2016.
- Pollack A (15 February 2017). "Harvard and M.I.T. Scientists Win Gene-Editing Patent Fight". The New York Times.
- Akst J (February 15, 2017). "Broad Wins CRISPR Patent Interference Case". The Scientist Magazine.
- Noonan KE (February 16, 2017). "PTAB Decides CRISPR Interference in Favor of Broad Institute -- Their Reasoning". Patent Docs.
- Potenza A (April 13, 2017). "UC Berkeley challenges decision that CRISPR patents belong to Broad Institute 3 comments The legal fight will likely continue for months or even years". The Verge. Retrieved 22 September 2017.
- Buhr S (July 26, 2017). "The CRISPR patent battle is back on as UC Berkeley files an appeal". TechCrunch. Retrieved 22 September 2017.
- Philippidis A (August 7, 2017). "MilliporeSigma to Be Granted European Patent for CRISPR Technology". Genetic Engineering & Biotechology News. Retrieved 22 September 2017.
- Akst J (March 24, 2017). "UC Berkeley Receives CRISPR Patent in Europe". The Scientist. Retrieved 22 September 2017.
- Cohen J (4 August 2017). "CRISPR patent battle in Europe takes a 'wild' twist with surprising player". Science. doi:10.1126/science.aan7211.
- "Top EU court: GMO rules cover plant gene editing technique". Retuers. 25 July 2018.
- AFP. "US Trial Shows 3 Cancer Patients Had Their Genomes Altered Safely by CRISPR". ScienceAlert. Retrieved 2020-02-09.
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (November 2013). "Genome engineering using the CRISPR-Cas9 system". Nature Protocols. 8 (11): 2281–308. doi:10.1038/nprot.2013.143. hdl:1721.1/102943. PMC 3969860. PMID 24157548.
- Ly J (2013). Discovering Genes Responsible for Kidney Diseases (Ph.D.). University of Toronto. Retrieved 26 December 2016.
- Mohr SE, Hu Y, Ewen-Campen B, Housden BE, Viswanatha R, Perrimon N (September 2016). "CRISPR guide RNA design for research applications". The FEBS Journal. 283 (17): 3232–8. doi:10.1111/febs.13777. PMC 5014588. PMID 27276584.
- Brazelton VA, Zarecor S, Wright DA, Wang Y, Liu J, Chen K, Yang B, Lawrence-Dill CJ (2015). "A quick guide to CRISPR sgRNA design tools". GM Crops & Food. 6 (4): 266–76. doi:10.1080/21645698.2015.1137690. PMC 5033207. PMID 26745836.
- Horvath P, Barrangou R (January 2010). "CRISPR/Cas, the immune system of bacteria and archaea". Science. 327 (5962): 167–70. Bibcode:2010Sci...327..167H. doi:10.1126/science.1179555. PMID 20056882. S2CID 17960960.
- Bialk P, Rivera-Torres N, Strouse B, Kmiec EB (2015-06-08). "Regulation of Gene Editing Activity Directed by Single-Stranded Oligonucleotides and CRISPR/Cas9 Systems". PLOS ONE. 10 (6): e0129308. Bibcode:2015PLoSO..1029308B. doi:10.1371/journal.pone.0129308. PMC 4459703. PMID 26053390.
- Sander JD, Joung JK (April 2014). "CRISPR-Cas systems for editing, regulating and targeting genomes". Nature Biotechnology. 32 (4): 347–55. doi:10.1038/nbt.2842. PMC 4022601. PMID 24584096.
- Lino CA, Harper JC, Carney JP, Timlin JA (November 2018). "Delivering CRISPR: a review of the challenges and approaches". Drug Delivery. 25 (1): 1234–1257. doi:10.1080/10717544.2018.1474964. PMC 6058482. PMID 29801422.
- Li L, Hu S, Chen X (July 2018). "Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities". Biomaterials. 171: 207–218. doi:10.1016/j.biomaterials.2018.04.031. PMC 5944364. PMID 29704747.
- Bak RO, Porteus MH (July 2017). "CRISPR-Mediated Integration of Large Gene Cassettes Using AAV Donor Vectors". Cell Reports. 20 (3): 750–756. doi:10.1016/j.celrep.2017.06.064. PMC 5568673. PMID 28723575.
- Schmidt F, Grimm D (February 2015). "CRISPR genome engineering and viral gene delivery: a case of mutual attraction". Biotechnology Journal. 10 (2): 258–72. doi:10.1002/biot.201400529. PMID 25663455.
- Waxmonsky N (24 September 2015). "CRISPR 101: Mammalian Expression Systems and Delivery Methods". Retrieved 11 June 2018.
- Oakes BL, Nadler DC, Flamholz A, Fellmann C, Staahl BT, Doudna JA, Savage DF (June 2016). "Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch". Nature Biotechnology. 34 (6): 646–51. doi:10.1038/nbt.3528. PMC 4900928. PMID 27136077.
- Nuñez JK, Harrington LB, Doudna JA (March 2016). "Chemical and Biophysical Modulation of Cas9 for Tunable Genome Engineering". ACS Chemical Biology. 11 (3): 681–8. doi:10.1021/acschembio.5b01019. PMID 26857072.
- Zhou W, Deiters A (April 2016). "Conditional Control of CRISPR/Cas9 Function". Angewandte Chemie. 55 (18): 5394–9. doi:10.1002/anie.201511441. PMID 26996256.
- Polstein LR, Gersbach CA (March 2015). "A light-inducible CRISPR-Cas9 system for control of endogenous gene activation". Nature Chemical Biology. 11 (3): 198–200. doi:10.1038/nchembio.1753. PMC 4412021. PMID 25664691.
- Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M (February 2015). "CRISPR-Cas9-based photoactivatable transcription system". Chemistry & Biology. 22 (2): 169–74. doi:10.1016/j.chembiol.2014.12.011. PMID 25619936.
- Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA (March 2015). "Rational design of a split-Cas9 enzyme complex". Proceedings of the National Academy of Sciences of the United States of America. 112 (10): 2984–9. Bibcode:2015PNAS..112.2984W. doi:10.1073/pnas.1501698112. PMC 4364227. PMID 25713377.
- Nihongaki Y, Kawano F, Nakajima T, Sato M (July 2015). "Photoactivatable CRISPR-Cas9 for optogenetic genome editing". Nature Biotechnology. 33 (7): 755–60. doi:10.1038/nbt.3245. PMID 26076431.
- Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A (May 2015). "Optical Control of CRISPR/Cas9 Gene Editing". Journal of the American Chemical Society. 137 (17): 5642–5. doi:10.1021/ja512664v. PMC 4919123. PMID 25905628.
- Jain PK, Ramanan V, Schepers AG, Dalvie NS, Panda A, Fleming HE, Bhatia SN (September 2016). "Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors". Angewandte Chemie. 55 (40): 12440–4. doi:10.1002/anie.201606123. PMC 5864249. PMID 27554600.
- Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR (May 2015). "Small molecule-triggered Cas9 protein with improved genome-editing specificity". Nature Chemical Biology. 11 (5): 316–8. doi:10.1038/nchembio.1793. PMC 4402137. PMID 25848930.
- Liu KI, Ramli MN, Woo CW, Wang Y, Zhao T, Zhang X, Yim GR, Chong BY, Gowher A, Chua MZ, Jung J, Lee JH, Tan MH (November 2016). "A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing". Nature Chemical Biology. 12 (11): 980–987. doi:10.1038/nchembio.2179. PMID 27618190. S2CID 33891039.
- Truong DJ, Kühner K, Kühn R, Werfel S, Engelhardt S, Wurst W, Ortiz O (July 2015). "Development of an intein-mediated split-Cas9 system for gene therapy". Nucleic Acids Research. 43 (13): 6450–8. doi:10.1093/nar/gkv601. PMC 4513872. PMID 26082496.
- Zetsche B, Volz SE, Zhang F (February 2015). "A split-Cas9 architecture for inducible genome editing and transcription modulation". Nature Biotechnology. 33 (2): 139–42. doi:10.1038/nbt.3149. PMC 4503468. PMID 25643054.
- González F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D (August 2014). "An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells". Cell Stem Cell. 15 (2): 215–26. doi:10.1016/j.stem.2014.05.018. PMC 4127112. PMID 24931489.
- Dow LE, Fisher J, O'Rourke KP, Muley A, Kastenhuber ER, Livshits G, Tschaharganeh DF, Socci ND, Lowe SW (April 2015). "Inducible in vivo genome editing with CRISPR-Cas9". Nature Biotechnology. 33 (4): 390–4. doi:10.1038/nbt.3155. PMC 4390466. PMID 25690852.
- Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, Qi LS (February 2015). "Small molecules enhance CRISPR genome editing in pluripotent stem cells". Cell Stem Cell. 16 (2): 142–7. doi:10.1016/j.stem.2015.01.003. PMC 4461869. PMID 25658371.
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL (May 2015). "Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining". Nature Biotechnology. 33 (5): 538–42. doi:10.1038/nbt.3190. PMC 4618510. PMID 25798939.
- Kurata M, Yamamoto K, Moriarity BS, Kitagawa M, Largaespada DA (February 2018). "CRISPR/Cas9 library screening for drug target discovery". Journal of Human Genetics. 63 (2): 179–186. doi:10.1038/s10038-017-0376-9. PMID 29158600.
- Hiranniramol K, Chen Y, Liu W, Wang X (January 2020). "Generalizable sgRNA design for improved CRISPR/Cas9 editing efficiency". Bioinformatics. 36 (9): 2684–2689. doi:10.1093/bioinformatics/btaa041. PMC 7203743. PMID 31971562.
- Agrotis A, Ketteler R (2015-09-24). "A new age in functional genomics using CRISPR/Cas9 in arrayed library screening". Frontiers in Genetics. 6: 300. doi:10.3389/fgene.2015.00300. PMC 4585242. PMID 26442115.
- Yu JS, Yusa K (July 2019). "Genome-wide CRISPR-Cas9 screening in mammalian cells". Methods. 164–165: 29–35. doi:10.1016/j.ymeth.2019.04.015. PMID 31034882.
- Stitzel M (2017-09-05). "Faculty of 1000 evaluation for Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening". Nature Protocols. 12 (4). doi:10.3410/f.727428415.793536376.
- Lin H, Jing H (2014-02-15). "Faculty of 1000 evaluation for Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library". doi:10.3410/f.718276719.793490932. Cite journal requires
|journal=(help) - "Addgene: Pooled Libraries". www.addgene.org. Retrieved 2020-01-31.
- McDade JR, Waxmonsky NC, Swanson LE, Fan M (July 2016). "Practical Considerations for Using Pooled Lentiviral CRISPR Libraries". Current Protocols in Molecular Biology. 115 (1): 31.5.1–31.5.13. doi:10.1002/cpmb.8. PMID 27366891.
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, et al. (October 2013). "Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system". Cell Research. 23 (10): 1163–71. doi:10.1038/cr.2013.122. PMC 3790238. PMID 23979020.
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. (October 2014). "Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation". Cell. 159 (3): 647–61. doi:10.1016/j.cell.2014.09.029. PMC 4253859. PMID 25307932.
- Dow LE (October 2015). "Modeling Disease In Vivo With CRISPR/Cas9". Trends in Molecular Medicine. 21 (10): 609–621. doi:10.1016/j.molmed.2015.07.006. PMC 4592741. PMID 26432018.
- Doudna J, Mali P (2016). CRISPR-Cas : a laboratory manual. Cold Spring Harbor, New York. ISBN 9781621821304. OCLC 922914104.
- Zuo E, Huo X, Yao X, Hu X, Sun Y, Yin J, et al. (2017). "CRISPR/Cas9-mediated targeted chromosome elimination". Genome Biology. 18 (1): 224. doi:10.1186/s13059-017-1354-4. PMC 5701507. PMID 29178945. Lay summary – Genome Web.
- Javed MR, Sadaf M, Ahmed T, Jamil A, Nawaz M, Abbas H, Ijaz A (August 2018). "CRISPR-Cas System: History and Prospects as a Genome Editing Tool in Microorganisms". review. Current Microbiology. 75 (12): 1675–1683. doi:10.1007/s00284-018-1547-4. PMID 30078067.
- Giersch RM, Finnigan GC (December 2017). "Yeast Still a Beast: Diverse Applications of CRISPR/Cas Editing Technology in S. cerevisiae". The Yale Journal of Biology and Medicine. 90 (4): 643–651. PMC 5733842. PMID 29259528.
- Raschmanová H, Weninger A, Glieder A, Kovar K, Vogl T (2018). "Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: Current state and future prospects". review. Biotechnology Advances. 36 (3): 641–665. doi:10.1016/j.biotechadv.2018.01.006. PMID 29331410.
- Ma D, Liu F (December 2015). "Genome Editing and Its Applications in Model Organisms". review. Genomics, Proteomics & Bioinformatics. 13 (6): 336–44. doi:10.1016/j.gpb.2015.12.001. PMC 4747648. PMID 26762955.
- Khurshid H, Jan SA, Shinwari ZK, Jamal M, Shah SH (2018). "An Era of CRISPR/ Cas9 Mediated Plant Genome Editing". review. Current Issues in Molecular Biology. 26: 47–54. doi:10.21775/cimb.026.047. PMID 28879855.
- Simone BW, Martínez-Gálvez G, WareJoncas Z, Ekker SC (August 2018). "Fishing for understanding: Unlocking the zebrafish gene editor's toolbox". review. Methods. 150: 3–10. doi:10.1016/j.ymeth.2018.07.012. PMC 6590056. PMID 30076892.
- Singh P, Schimenti JC, Bolcun-Filas E (January 2015). "A mouse geneticist's practical guide to CRISPR applications". review. Genetics. 199 (1): 1–15. doi:10.1534/genetics.114.169771. PMC 4286675. PMID 25271304.
- Soni D, Wang DM, Regmi SC, Mittal M, Vogel SM, Schlüter D, Tiruppathi C (May 2018). "Deubiquitinase function of A20 maintains and repairs endothelial barrier after lung vascular injury". Cell Death Discovery. 4 (60): 60. doi:10.1038/s41420-018-0056-3. PMC 5955943. PMID 29796309.
- Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B, et al. (2018). "Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents". Nature. 553 (7687): 217–221. Bibcode:2018Natur.553..217G. doi:10.1038/nature25164. PMC 5784267. PMID 29258297.
- Kadam US, Shelake RM, Chavhan RL, Suprasanna P (October 2018). "Concerns regarding 'off-target' activity of genome editing endonucleases". review. Plant Physiology and Biochemistry. 131: 22–30. doi:10.1016/j.plaphy.2018.03.027. PMID 29653762.
- Kimberland ML, Hou W, Alfonso-Pecchio A, Wilson S, Rao Y, Zhang S, Lu Q (August 2018). "Strategies for controlling CRISPR/Cas9 off-target effects and biological variations in mammalian genome editing experiments". review. Journal of Biotechnology. 284: 91–101. doi:10.1016/j.jbiotec.2018.08.007. PMID 30142414.
- van Erp PB, Bloomer G, Wilkinson R, Wiedenheft B (June 2015). "The history and market impact of CRISPR RNA-guided nucleases". Current Opinion in Virology. 12: 85–90. doi:10.1016/j.coviro.2015.03.011. PMC 4470805. PMID 25914022.
- Maggio I, Gonçalves MA (May 2015). "Genome editing at the crossroads of delivery, specificity, and fidelity". Trends in Biotechnology. 33 (5): 280–91. doi:10.1016/j.tibtech.2015.02.011. PMID 25819765.
- Rath D, Amlinger L, Rath A, Lundgren M (October 2015). "The CRISPR-Cas immune system: biology, mechanisms and applications". Biochimie. 117: 119–28. doi:10.1016/j.biochi.2015.03.025. PMID 25868999.
- "What Is CRISPR? How Does It Work? Is It Gene Editing? » LiveScience.Tech". LiveScience.Tech. 2018-04-30. Retrieved 2020-02-06.
- Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, et al. (October 2015). "Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids". Nature Communications. 6: 8715. Bibcode:2015NatCo...6.8715F. doi:10.1038/ncomms9715. PMC 4620584. PMID 26493500.
- Cruz NM, Song X, Czerniecki SM, Gulieva RE, Churchill AJ, Kim YK, et al. (November 2017). "Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease". Nature Materials. 16 (11): 1112–1119. Bibcode:2017NatMa..16.1112C. doi:10.1038/nmat4994. PMC 5936694. PMID 28967916.
- Kim YK, Refaeli I, Brooks CR, Jing P, Gulieva RE, Hughes MR, et al. (December 2017). "Gene-Edited Human Kidney Organoids Reveal Mechanisms of Disease in Podocyte Development". Stem Cells. 35 (12): 2366–2378. doi:10.1002/stem.2707. PMC 5742857. PMID 28905451.
- Bellin M, Casini S, Davis RP, D'Aniello C, Haas J, Ward-van Oostwaard D, Tertoolen LG, Jung CB, Elliott DA, Welling A, Laugwitz KL, Moretti A, Mummery CL (December 2013). "Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome". The EMBO Journal. 32 (24): 3161–75. doi:10.1038/emboj.2013.240. PMC 3981141. PMID 24213244.
- Cai L, Fisher AL, Huang H, Xie Z (December 2016). "CRISPR-mediated genome editing and human diseases". Genes & Diseases. 3 (4): 244–251. doi:10.1016/j.gendis.2016.07.003. PMC 6150104. PMID 30258895.
- "Seven Diseases That CRISPR Technology Could Cure". Labiotech.eu. 2018-06-25. Retrieved 2018-08-22.
- "CRISPR/Cas9 and Cancer". Immuno-Oncology News. 2018-04-27. Retrieved 2019-02-18.
- Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW (September 2014). "Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac". Genome Research. 24 (9): 1526–33. doi:10.1101/gr.173427.114. PMC 4158758. PMID 25096406.
- Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, Uchida N, Hendel A, Narla A, Majeti R, Weinberg KI, Porteus MH (2016-11-17). "CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells". Nature. 539 (7629): 384–389. Bibcode:2016Natur.539..384D. doi:10.1038/nature20134. PMC 5898607. PMID 27820943.
- "CRISPR "One Shot Cell Therapy for Hemophilia Developed | GEN". GEN. 2018-05-02. Retrieved 2018-08-22.
- Marangi M, Pistritto G (2018-04-20). "Innovative Therapeutic Strategies for Cystic Fibrosis: Moving Forward to CRISPR Technique". Frontiers in Pharmacology. 9: 396. doi:10.3389/fphar.2018.00396. PMC 5920621. PMID 29731717.
- Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS (February 2017). "Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy". Nature Communications. 8: 14454. Bibcode:2017NatCo...814454B. doi:10.1038/ncomms14454. PMC 5316861. PMID 28195574.
- Eisenstein M (May 2018). "CRISPR takes on Huntington's disease". Nature. 557 (7707): S42–S43. Bibcode:2018Natur.557S..42E. doi:10.1038/d41586-018-05177-y. PMID 29844549.
- Dabrowska M, Juzwa W, Krzyzosiak WJ, Olejniczak M (2018). "Precise Excision of the CAG Tract from the Huntingtin Gene by Cas9 Nickases". Frontiers in Neuroscience. 12: 75. doi:10.3389/fnins.2018.00075. PMC 5834764. PMID 29535594.
- King A (March 2018). "A CRISPR edit for heart disease". Nature. 555 (7695): S23–S25. Bibcode:2018Natur.555.....K. doi:10.1038/d41586-018-02482-4. PMID 29517035.
- Abrahimi P, Chang WG, Kluger MS, Qyang Y, Tellides G, Saltzman WM, Pober JS (July 2015). "Efficient gene disruption in cultured primary human endothelial cells by CRISPR/Cas9". Circulation Research. 117 (2): 121–8. doi:10.1161/CIRCRESAHA.117.306290. PMC 4490936. PMID 25940550.
- Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL (January 2014). "Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems". mBio. 5 (1): e00928–13. doi:10.1128/mBio.00928-13. PMC 3903277. PMID 24473129.
- Citorik RJ, Mimee M, Lu TK (November 2014). "Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases". Nature Biotechnology. 32 (11): 1141–5. doi:10.1038/nbt.3011. hdl:1721.1/100834. PMC 4237163. PMID 25240928.
- Gholizadeh P, Aghazadeh M, Asgharzadeh M, Kafil HS (October 2017). "Suppressing the CRISPR/Cas adaptive immune system in bacterial infections". European Journal of Clinical Microbiology & Infectious Diseases. 36 (11): 2043–2051. doi:10.1007/s10096-017-3036-2. PMID 28601970.
- Gibney, Elizabeth (January 2, 2018). "What to expect in 2018: science in the new year". Nature. 553 (7686): 12–13. Bibcode:2018Natur.553...12G. doi:10.1038/d41586-018-00009-5. PMID 29300040.
- Taylor, Phil (Jan 3, 2019). "J&J takes stake in Locus' CRISPR-based 'Pac-Man' antimicrobials". Fierce Biotech. Retrieved 27 February 2019.
- Reardon, Sara (2017). "Modified viruses deliver death to antibiotic-resistant bacteria". Nature. 546 (7660): 586–587. Bibcode:2017Natur.546..586R. doi:10.1038/nature.2017.22173. PMID 28661508.
- van Diemen FR, Kruse EM, Hooykaas MJ, Bruggeling CE, Schürch AC, van Ham PM, Imhof SM, Nijhuis M, Wiertz EJ, Lebbink RJ (2016). "CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections". PLOS Pathogens. 12 (6): e1005701. doi:10.1371/journal.ppat.1005701. PMC 4928872. PMID 27362483. Lay summary – PLOS Media YouTube Channel.
- Science News Staff (December 17, 2015). "And Science's Breakthrough of the Year is …". news.sciencemag.org. Retrieved 2015-12-21.
- Mullin E. "Using CRISPR on pigs could make their organs safer for human transplant". MIT Technology Review. Retrieved 2017-09-09.
- Khan FA, Pandupuspitasari NS, Chun-Jie H, Ao Z, Jamal M, Zohaib A, Khan FA, Hakim MR, ShuJun Z (August 2016). "CRISPR/Cas9 therapeutics: a cure for cancer and other genetic diseases". Oncotarget. 7 (32): 52541–52552. doi:10.18632/oncotarget.9646. PMC 5239572. PMID 27250031.
- Reardon S (2016). "First CRISPR clinical trial gets green light from US panel". Nature. doi:10.1038/nature.2016.20137.
- Dominguez AA, Lim WA, Qi LS (January 2016). "Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation". Nature Reviews Molecular Cell Biology. 17 (1): 5–15. doi:10.1038/nrm.2015.2. PMC 4922510. PMID 26670017.
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F (January 2014). "Genome-scale CRISPR-Cas9 knockout screening in human cells". Science. 343 (6166): 84–7. Bibcode:2014Sci...343...84S. doi:10.1126/science.1247005. PMC 4089965. PMID 24336571.
- Pennisi E (August 2013). "The CRISPR craze". News Focus. Science. 341 (6148): 833–6. Bibcode:2013Sci...341..833P. doi:10.1126/science.341.6148.833. PMID 23970676.
- Zimmer C (2016-06-03). "Scientists Find Form of Crispr Gene Editing With New Capabilities". The New York Times. ISSN 0362-4331. Retrieved 2016-06-10.
- Pickar-Oliver, Adrian; Gersbach, Charles A. (2019). "The next generation of CRISPR–Cas technologies and applications". Nature Reviews Molecular Cell Biology. 20 (8): 490–507. doi:10.1038/s41580-019-0131-5. ISSN 1471-0072. PMC 7079207. PMID 31147612.
- Basak J, Nithin C (2015). "Targeting Non-Coding RNAs in Plants with the CRISPR-Cas Technology is a Challenge yet Worth Accepting". Frontiers in Plant Science. 6: 1001. doi:10.3389/fpls.2015.01001. PMC 4652605. PMID 26635829.
- Akbari OS, Bellen HJ, Bier E, Bullock SL, Burt A, Church GM, et al. (August 2015). "BIOSAFETY. Safeguarding gene drive experiments in the laboratory". Science. 349 (6251): 927–9. Bibcode:2015Sci...349..927A. doi:10.1126/science.aac7932. PMC 4692367. PMID 26229113.
- Caplan AL, Parent B, Shen M, Plunkett C (November 2015). "No time to waste--the ethical challenges created by CRISPR: CRISPR/Cas, being an efficient, simple, and cheap technology to edit the genome of any organism, raises many ethical and regulatory issues beyond the use to manipulate human germ line cells". EMBO Reports. 16 (11): 1421–6. doi:10.15252/embr.201541337. PMC 4641494. PMID 26450575.
- Oye KA, Esvelt K, Appleton E, Catteruccia F, Church G, Kuiken T, et al. (August 2014). "Biotechnology. Regulating gene drives". Science. 345 (6197): 626–8. Bibcode:2014Sci...345..626O. doi:10.1126/science.1254287. PMID 25035410.
- Gu W, Crawford ED, O'Donovan BD, Wilson MR, Chow ED, Retallack H, DeRisi JL (March 2016). "Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications". Genome Biology. 17: 41. doi:10.1186/s13059-016-0904-5. PMC 4778327. PMID 26944702.
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR (2019). "Search-and-replace genome editing without double-strand breaks or donor DNA". Nature. 576 (7785): 149–157. Bibcode:2019Natur.576..149A. doi:10.1038/s41586-019-1711-4. PMC 6907074. PMID 31634902.
- A New Gene Editing Tool Could Make CRISPR More Precise. Lila Thulin, The Smithsonian Magazine. 21 October 2019.
- New 'prime' genome editor could surpass CRISPR. Jon Cohen, Science. 21 October 2019.
- New "Prime Editing" Method Makes Only Single-Stranded DNA Cuts. Emma Yasinski, The Scientist. 21 October 2019.
- Prime editing: DNA tool could correct 89% of genetic defects. James Gallagher, BBC News. 21 October 2019.
- Regalado A (March 5, 2015). "Engineering the Perfect Baby". MIT Technology Review.
- Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, Corn JE, Daley GQ, Doudna JA, Fenner M, Greely HT, Jinek M, Martin GS, Penhoet E, Puck J, Sternberg SH, Weissman JS, Yamamoto KR (April 2015). "Biotechnology. A prudent path forward for genomic engineering and germline gene modification". Science. 348 (6230): 36–8. Bibcode:2015Sci...348...36B. doi:10.1126/science.aab1028. PMC 4394183. PMID 25791083.
- Lanphier E, Urnov F, Haecker SE, Werner M, Smolenski J (March 2015). "Don't edit the human germ line". Nature. 519 (7544): 410–1. Bibcode:2015Natur.519..410L. doi:10.1038/519410a. PMID 25810189.
- Wade N (19 March 2015). "Scientists Seek Ban on Method of Editing the Human Genome". The New York Times. Retrieved 20 March 2015.
The biologists writing in Science support continuing laboratory research with the technique, and few if any scientists believe it is ready for clinical use.
- Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, Huang J (May 2015). "CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes". Protein & Cell. 6 (5): 363–72. doi:10.1007/s13238-015-0153-5. PMC 4417674. PMID 25894090.
- Kolata G (23 April 2015). "Chinese Scientists Edit Genes of Human Embryos, Raising Concerns". The New York Times. Retrieved 24 April 2015.
- Cyranoski D, Reardon S (2015). "Chinese scientists genetically modify human embryos". Nature. doi:10.1038/nature.2015.17378.
- Regalado A (2016-05-08). "Chinese Researchers Experiment with Making HIV-Proof Embryos". MIT Technology Review. Retrieved 2016-06-10.
- "International Summit on Gene Editing". National Academies of Sciences, Engineering, and Medicine. 3 December 2015. Retrieved 3 December 2015.
- Brokowski, Carolyn (April 2018). "Do CRISPR Germline Ethics Statements Cut It?". CRISPR Journal. 1 (2): 115–125. doi:10.1089/crispr.2017.0024. PMC 6694771. PMID 31021208.
- Begley S (28 November 2018). "Amid uproar, Chinese scientist defends creating gene-edited babies". STAT.
- editor, Ian Sample Science (13 March 2019). "Scientists call for global moratorium on gene editing of embryos". Theguardian.com. Retrieved 14 March 2019.CS1 maint: extra text: authors list (link)
- Callaway E (February 2016). "UK scientists gain licence to edit genes in human embryos". Nature. 530 (7588): 18. Bibcode:2016Natur.530...18C. doi:10.1038/nature.2016.19270. PMID 26842037.
- McHughen A, Smyth S (January 2008). "US regulatory system for genetically modified [genetically modified organism (GMO), rDNA or transgenic] crop cultivars". Plant Biotechnology Journal. 6 (1): 2–12. doi:10.1111/j.1467-7652.2007.00300.x. PMID 17956539. S2CID 3210837.
- USDA. "Re: Request to confirm" (PDF).
- Waltz E (2016). "Gene-edited CRISPR mushroom escapes US regulation". Nature. 532 (7599): 293. Bibcode:2016Natur.532..293W. doi:10.1038/nature.2016.19754. PMID 27111611.
- Ledford H (April 2016). "Gene-editing surges as US rethinks regulations". Nature. 532 (7598): 158–9. Bibcode:2016Natur.532..158L. doi:10.1038/532158a. PMID 27075074.
- "The FDA Is Cracking Down On Rogue Genetic Engineers", Kristen V. Brown. Gizmodo. February 1, 2017. Retrieved 5 feb 2017
- "Guidance for Industry #187 / Regulation of Intentionally Altered Genomic DNA in Animals" (PDF). 2020-02-11.
- Cyranoski D (2017). "China's embrace of embryo selection raises thorny questions". Nature. 548 (7667): 272–274. Bibcode:2017Natur.548..272C. doi:10.1038/548272a. PMID 28816265.
- Peng Y (2016). "The morality and ethics governing CRISPR-Cas9 patents in China". Nature Biotechnology. 34 (6): 616–8. doi:10.1038/nbt.3590. PMID 27281418.
- Rana P, Marcus AD, Fan W (2018-01-21). "China, Unhampered by Rules, Races Ahead in Gene-Editing Trials". Wall Street Journal. ISSN 0099-9660. Retrieved 2018-01-23.
- Talbot D (2016). "Precise Gene Editing in Plants/ 10 Breakthrough Technologies 2016". MIT Technology review. Massachusetts Institute of Technology. Retrieved 18 March 2016.
- Larson C, Schaffer A (2014). "Genome Editing/ 10 Breakthrough Technologies 2014". Massachusetts Institute of Technology. Retrieved 18 March 2016.
- 良艮創意, 很好設計, 李維宗設計. "Tang Prize Laureates". www.tang-prize.org. Retrieved 2018-08-05.