Transfer DNA
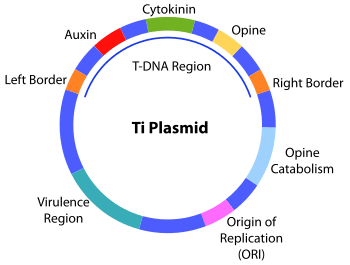
The transfer DNA (abbreviated T-DNA) is the transferred DNA of the tumor-inducing (Ti) plasmid of some species of bacteria such as Agrobacterium tumefaciens and [[Agrobacterium rhizogenes](actually an Ri plasmid)]. The T-DNA is transferred from bacterium into the host plant's nuclear DNA genome. The capability of this specialized tumor-inducing (Ti) plasmid is attributed to two essential regions required for DNA transfer to the host cell. As the T-DNA is bordered by 25-base-pair repeats on each end. Transfer is initiated at the right border and terminated at the left border and requires the vir genes of the Ti plasmid.
The bacterial T-DNA is about 24,000 base pairs long[1][2] and contains genes that code for enzymes synthesizing opines and phytohormones. By transferring the T-DNA into the plant genome, the bacterium essentially reprograms the plant cells to grow into a tumor and produce a unique food source for the bacteria. The synthesis of the plant hormones auxin and cytokinin by enzymes encoded in the T-DNA enables the plant cell to grow uncontrollably, thus forming the crown gall tumors typically induced by Agrobacterium tumefaciens infection.[3] Whereas Agrobacterium rhizogenes causes hairy root disease. The opines are amino acid derivatives used by the bacterium as a source of carbon and energy. This natural process of horizontal gene transfer in plants is being utilized as a tool for fundamental and applied research in plant biology through Agrobacterium tumefaciens mediated foreign gene transformation and insertional mutagenesis.[4][5]
Transformation
Agrobacterium-mediated T-DNA transfer is widely used as a tool in biotechnology. For more than two decades, Agrobacterium tumefaciens has been exploited for introducing genes into plants for basic research as well as for commercial production of transgenic crops.[6] In genetic engineering, the tumor-promoting and opine-synthesis genes are removed from the T-DNA and replaced with a gene of interest and/or a selection marker, which is required to establish which plants have been successfully transformed. Examples of selection markers include neomycin phosphotransferase, hygromycin B phosphotransferase (which both phosphorylate antibiotics) and phosphinothricin acetyltransferase (which acetylates and deactivates phosphinothricin, a potent inhibitor of glutamine synthetase) or a herbicide formulations such as Basta or Bialophos.[7] Another selection system that can be employed is usage of metabolic markers such as phospho-mannose isomerase.[8] Agrobacterium is then used as a vector to transfer the engineered T-DNA into the plant cells where it integrates into the plant genome. This method can be used to generate transgenic plants carrying a foreign gene. Agrobacterium tumefaciens is capable of transferring foreign DNA to both monocotyledons and dicotyledonous plants efficiently while taking care of critically important factors like the genotype of plants, types and ages of tissues inoculated, kind of vectors, strains of Agrobacterium, selection marker genes and selective agents, and various conditions of tissue culture.[3]
Mechanism
The infection process of T-DNA into the host cell and integration into its nucleus involve multiple steps. First, the bacteria multiply in the wound sap before infection and then attach to the plant cell walls. The bacterial virulence genes expression of approximately 10 operons is activated by perception of phenolic compounds such as acetosyringone emitted by wounded plant tissue and follows cell-cell contact. Then this process succeeds the macromolecular translocation from Agrobacterium to cytoplasm of host cell, transmission of T-DNA along with associated proteins (called T-complex) to the host cell nucleus followed by disassembly of the T-complex, stable integration of T-DNA into host plant genome, and expression of the transferred genes. The integration of T-DNA into a host genome involves the formation of a nick at the right border of the Ti plasmid. This nick creates a region of single stranded DNA from the left border of the T-DNA gene over to the right border which was cut. Then, single stranded binding proteins attach to the single stranded DNA. DNA synthesis displaces the single stranded region and then a second nick at the left border region releases the single stranded T-DNA fragment. Further this fragment can be incorporated into a host genome.[9]
Agrobacterium has been known to evolve a control system that hijacks host factors and cellular processes for several pathways of host-plant defense response to invade the host cell nucleus. For the integration of T-DNA into the target host genome, Agrobacterium carries out multiple interactions with host-plant factors.[9] To interact with host plant proteins many Agrobacterium virulence proteins encoded by vir genes. Agrobacterium vir genes expression occurs via the VirA-VirG sensor that results in generation of a mobile single-stranded T-DNA copy (T-strand). Processed form of VirB2 is the major component of the T-complex that is required for transformation. VirD2 is the protein that caps the 5′ end of the transferred T-strand by covalent attachment and is transported to the host cell cytoplasm.[10] VirE2 is the single-stranded DNA binding protein that presumably coats the T- strand in the host cytoplasm by cooperative binding. It is then directed into the nucleus via interactions with the host cell proteins such as importin a, bacterial VirE3, and dynein-like proteins. Several other bacterial virulence effectors like VirB5, VirB7 (the minor components of the T-complex), VirD5, VirE2, VirE3, and VirF that may also interact with proteins of host plant cells.[11]
Uses in mutagenesis
The same procedure of T-DNA transfer can be used to disrupt genes via insertional mutagenesis.[5] Not only does the inserted T-DNA sequence create a mutation but it also 'tags'[12] the affected gene, thus allowing for its isolation. A reporter gene can be linked to the right end of the T-DNA to be transformed along with a plasmid replicon and a selectable antibiotic (such as hygromycin)-resistance gene and can explicit approximately 30% of average efficiency having successful T-DNA inserts induced gene fusions in Arabidopsis thaliana.[13]
Reverse genetics is usually followed as a functional genomics approach based on the dynamic of biological system that aims to assign a function(s) to genes and determines how genes and their products interact together. Transgenics involving screening of populations by T-DNA insertional mutagenesis is one of the powerful to study loss of function and ectopic expression to assign that function to the gene under investigation.[14] Hence, this method is used widely to study gene function in plants, such as the model plant Arabidopsis thaliana. While studying T-DNA insertion for gene disruption Arabidopsis thaliana mutants may also show different phenotypic classes like seedling-lethals, size variants, pigment, embryo-defective, reduced-fertility, dramatic (morphological), and physiological.[15] For example, this gene disruption strategy used for assigning functions to genes defined only by sequence helped to demonstrate En-1 insertion in the flavanone 3-hydroxylase gene (F3H). Knock-out alleles of En-1 insertion in the flavonol synthase gene (FLS) were characterized by this reverse genetics approach that drastically reduced levels of kaempferol.[16]
See also
References
- ↑ Barker, R. F.; Idler, K. B.; Thompson, D. V.; Kemp, J. D. (1983-11-01). "Nucleotide sequence of the T-DNA region from theA grobacterium tumefaciens octopine Ti plasmid pTi15955". Plant Molecular Biology. 2 (6): 335–350. doi:10.1007/BF01578595. ISSN 0167-4412.
- ↑ Gielen, J.; Terryn, N.; Villarroel, R.; Van Montagu, M. (1999-08-01). "Complete nucleotide sequence of the T-DNA region of the plant tumour-inducing Agrobacterium tumefaciens Ti plasmid pTiC58". Journal of Experimental Botany. 50 (337): 1421–1422. doi:10.1093/jxb/50.337.1421. ISSN 0022-0957.
- 1 2 Hiei, Yukoh; Komari, Toshihiko; Kubo, Tomoaki (1997-09-01). "Transformation of rice mediated by Agrobacterium tumefaciens". Plant Molecular Biology. 35 (1–2): 205–218. doi:10.1023/a:1005847615493. ISSN 0167-4412.
- ↑ Zupan, J R; Zambryski, P (April 1995). "Transfer of T-DNA from Agrobacterium to the plant cell". Plant Physiology. 107 (4): 1041–1047. doi:10.1104/pp.107.4.1041. ISSN 0032-0889. PMC 157234. PMID 7770515.
- 1 2 Krysan PJ, Young JC, Sussman MR (December 1999). "T-DNA as an insertional mutagen in Arabidopsis". The Plant Cell. 11 (12): 2283–90. doi:10.1105/tpc.11.12.2283. ISSN 1040-4651. PMC 144136. PMID 10590158.
- ↑ Oltmanns, Heiko; Frame, Bronwyn; Lee, Lan-Ying; Johnson, Susan; Li, Bo; Wang, Kan; Gelvin, Stanton B. (2010-03-01). "Generation of Backbone-Free, Low Transgene Copy Plants by Launching T-DNA from the Agrobacterium Chromosome". Plant Physiology. 152 (3): 1158–1166. doi:10.1104/pp.109.148585. ISSN 0032-0889. PMC 2832237. PMID 20023148.
- ↑ Lee LY, Gelvin SB (February 2008). "T-DNA binary vectors and systems". Plant Physiology. 146 (2): 325–32. doi:10.1104/pp.107.113001. ISSN 0032-0889. PMC 2245830. PMID 18250230.
- ↑ Todd, Rebecca; Tague, Brian W. (2001-12-01). "Phosphomannose isomerase: A versatile selectable marker forArabidopsis thaliana germ-line transformation". Plant Molecular Biology Reporter. 19 (4): 307–319. doi:10.1007/bf02772829. ISSN 0735-9640.
- 1 2 Lacroix B, Citovsky V (2013). "The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation". The International Journal of Developmental Biology. 57 (6–8): 467–81. doi:10.1387/ijdb.130199bl. ISSN 0214-6282. PMID 24166430.
- ↑ Koukolíková-Nicola, Z; Raineri, D; Stephens, K; Ramos, C; Tinland, B; Nester, E W; Hohn, B (February 1993). "Genetic analysis of the virD operon of Agrobacterium tumefaciens: a search for functions involved in transport of T-DNA into the plant cell nucleus and in T-DNA integration". Journal of Bacteriology. 175 (3): 723–731. doi:10.1128/jb.175.3.723-731.1993. ISSN 0021-9193. PMC 196211. PMID 8380800.
- ↑ Gelvin, Stanton B. (2003-03-01). "Agrobacterium-Mediated Plant Transformation: the Biology behind the "Gene-Jockeying" Tool". Microbiology and Molecular Biology Reviews. 67 (1): 16–37. doi:10.1128/mmbr.67.1.16-37.2003. ISSN 1092-2172. PMC 150518. PMID 12626681.
- ↑ Liu, Yao-Guang; Shirano, Yumiko; Fukaki, Hidehiro; Yanai, Yukihiro; Tasaka, Masao; Tabata, Satoshi; Shibata, Daisuke (1999-05-25). "Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning". Proceedings of the National Academy of Sciences. 96 (11): 6535–6540. doi:10.1073/pnas.96.11.6535. ISSN 0027-8424. PMC 26917. PMID 10339623.
- ↑ Koncz C, Martini N, Mayerhofer R, Koncz-Kalman Z, Körber H, Redei GP, Schell J (November 1989). "High-frequency T-DNA-mediated gene tagging in plants". Proceedings of the National Academy of Sciences of the United States of America. 86 (21): 8467–71. doi:10.1073/pnas.86.21.8467. PMC 298303. PMID 2554318.
- ↑ Ben-Amar A, Daldoul S, Reustle GM, Krczal G, Mliki A (December 2016). "Reverse Genetics and High Throughput Sequencing Methodologies for Plant Functional Genomics". Current Genomics. 17 (6): 460–475. doi:10.2174/1389202917666160520102827. PMC 5282599. PMID 28217003.
- ↑ Feldmann, Kenneth A. (1991-07-01). "T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum". The Plant Journal. 1 (1): 71–82. doi:10.1111/j.1365-313x.1991.00071.x. ISSN 1365-313X.
- ↑ Wisman, Ellen; Hartmann, Ulrike; Sagasser, Martin; Baumann, Elvira; Palme, Klaus; Hahlbrock, Klaus; Saedler, Heinz; Weisshaar, Bernd (1998-10-13). "Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes". Proceedings of the National Academy of Sciences. 95 (21): 12432–12437. doi:10.1073/pnas.95.21.12432. ISSN 0027-8424. PMC 22848. PMID 9770503.
- P.H. Raven, R.F. Evert, S.E. Eichhorn (2005): Biology of Plants, 7th Edition, W.H. Freeman and Company Publishers, New York, ISBN 0-7167-1007-2