Phytoglobin
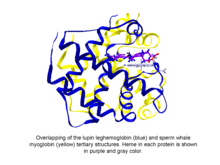
Phytoglobins are globular plant (algae and land plant) proteins classified into the globin superfamily, which contain a heme, i.e. protoporphyrin IX-Fe, prosthetic group. Phytoglobins were discovered in 1939 by Kubo after spectroscopic and chemical analysis of the red pigment of soybean root nodules.[1] A few decades after Kubo's report the crystallization of a lupin phytoglobin (known as leghemoglobin) by Vainshtein and collaborator revealed that the tertiary structure of this protein and that of the sperm whale myoglobin was remarkably similar, thus indicating that the phytoglobin discovered by Kubo did indeed correspond to a globin.[2]
Distribution and classification
Phytoglobins (abbreviated as Phytogbs) are ubiquitously distributed in plants as they have been identified in algae and land plants, including primitive bryophytes and evolved monocots and dicots. They are classified as Phytogbs0, Phytogbs1 and Phytogbs2 (formerly known as non-symbiotic hemoglobins), symbiotic Phytogbs, leghemoglobins (abbreviated as Lbs) and Phytogbs3.[3] The Phytogbs0 are primitive Phytogbs found in algae, bryophytes and gymnosperms. The Phytogbs1 and Phytogbs2 apparently evolved from Phytogbs0 and are found in angiosperms. Symbiotic Phytogbs (abbreviated as SymPhytogbs) and Lbs are specialized Phytogbs localized in nodules of nitrogen-fixing non-legumes, such as the actinorhizal plants that establish a symbiotic association with the actinomycete Frankia, and legumes that establish a symbiotic association with rhizobia bacteria, respectively. The Phytogbs3 are structurally similar to the bacterial truncated globins and are found in algae and land plants.
Structural characteristics
Phytogbs are coded by genes interrupted by 3 introns (although a 4 introns-containing phytogb gene has been detected in the moss Physcomitrella patens ).[4] The first and third intron of the phytogb genes are localized in the same position as that of the myoglobin genes, which suggests that phytogb and myoglobin genes evolved from a common ancestor more than 600 million years ago.[5] The existence of a second intron in the phytogb genes was predicted by Go using theoretical analysis,[6] which was further verified by cloning and sequencing of soybean lb genes by Marker and collaborators.[7]
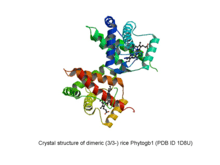
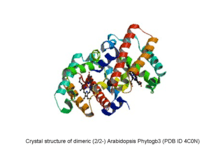
Phytogbs are monomeric proteins whose molecular mass ranges from ~17 to ~19 kDa. However, at high (more than 1 mM) concentrations Phytogbs can form dimers. Phytogbs polypeptide chain folds into a particular arrangement of 6 to 7 helices (named with letters A to H) known as the globin fold which forms a hydrophobic pocket where heme is located. Two types of the globin fold have been identified in Phytogbs: the 3/3- and 2/2-folding,[8][9] where helices A, E and F overlap to helices B, G and H and helices B and E overlap to helices G and H, respectively.
Like other globins, heme-Fe in Phytogbs is coordinated at the proximal region by a His amino acid (named as proximal His). The distal region of heme-Fe can be occupied by either a variety of ligands (such as oxygen and nitric oxide) or a distal (frequently a His) amino acid, giving place to penta- or hexa-coordinate Phytogbs, respectively. The Phytogbs2, SymPhytogbs and Lbs are predominantly penta-coordinate whereas Phytogbs1 are predominantly hexa-coordinate and Phytogbs0 and Phytogbs3 are a combination of penta- and hexa-coordinate.[3] Heme-Fe coordination is essential for Phytogb (and other globins) function because it regulates the rate of ligand-binding and –releasing as consequence of the kinetic constants kon and koff, respectively. For example, the affinity of soybean Lb and rice Phytogb1 for O2 (KO2) is moderate and very high because kon is 130 and 68 mM−1 s−1, koff is 5.6 and 0.038 s−1 and KO2 (i.e. the O2-affinity resulting from kon/koff) is 23 and 1800 mM−1, respectively.[10] This indicates that soybean Lb could function as an O2-storage or –transport protein and that the function of rice Phytogb1 (and other hexa-coordinate Phytogbs) could be other than O2-transport because the high affinity of this protein for O2 results from an extremely low koff constant.[11]
Synthesis and postulated functions
Like other globins, penta-coordinate Phytogbs reversibly bind and transport O2. The function of Lbs in nodules was elucidated in 1974 by Wittenberg, Appleby and others.[12] In nodules the concentration of Lbs is very high as they correspond to ~30% of the total soluble proteins. The apparent function of Lbs in nodules is to facilitate the diffusion of O2 to the respiring bacteroids for nitrogen-fixation. At the same time, Lb contributes to maintain low O2-levels (~10 nM) to avoid inactivation of the O2-sensitive nitrogenase that fixes the atmospheric nitrogen.[13]
Furthermore, Phytogbs bind other gaseous ligands, most notably nitric oxide (NO), and exhibit a NO dioxygenase activity.[14] Work by Hill and collaborators during the last ~15 years has shown that levels of endogenous NO varies with the concentration of Phytogbs1 in transgenic maize and alfalfa.[15] Based on these observations, these authors have proposed that a function of oxygenated Phytogbs is to modulate levels of NO via an NO dioxygenase activity and to indirectly regulate a wide variety of cell functions that are modulated by levels of NO.
Phytogbs0, 1, 2 and 3 are synthesized at very low concentrations in diverse (embryonic and vegetative) plant organs.[16][17][18] However, concentrations of Phytogbs increase in plants subjected to specific stress conditions, such as flooding[19] and light-limitation.[20] Hence, some Phytogbs have been considered as plant stress-responsive proteins.
Evolution
Work by Arredondo-Peter, Vinogradov and collaborators has elucidated the major events that could have occurred during the evolution of Phytogbs.[21][22][23] Although Phytogbs evolved from a common ancestor previous to the origin of eukaryotes (i.e. in the eubacteria domain), in plants two evolutionary lineages have been identified for Phytogbs: the (3/3-) Phytogb0, 1 and 2 and SymPhytogb and Lb lineage and the (2/2-) Phytogb3 lineage. Apparently, Phytogbs0 were the ancestors to Phytogbs1 and 2, and SymPhytogbs and Lbs evolved from a Phytogb1 ancestor as an adaptation to the symbiotic nitrogen-fixation. Comparative analysis of moss Phytogb0 with rice Phytogb1 and soybean Lb structure revealed that the major evolutionary changes that probably occurred during the evolution of Phytogbs0, 1 and 2, SymPhytogbs and Lbs were (i) a hexa-coordinate to penta-coordinate transition at the heme group, (ii) a length decrease at the CD-loop and N- and C-termini, and (iii) the compaction of the protein into a globular structure. Furthermore, the structure of the globin domain from Phytogbs3 is highly similar to that of bacterial truncated globins indicating that the 2/2-folding conserved during the evolution of Phytogbs3. However, globin domain from land plant Phytogbs3 is flanked by extra-amino acid sequences that probably originated in the ancestor to land plant Phytogbs3. The functional and evolutionary significance of the extra-amino acid sequences in the Phytogbs3 structure is still not known.
Technological applications
Phytogbs are potentially useful for a number of biotechnological applications. For example, shortly after the discovery of rice Phytogbs1 an intended application was the use of these proteins as O2-sensors in electronic devices. Also, over-expression of Phytogbs has been proposed as a strategy to increase crop tolerance to specific stress conditions, such as crop flooding. Recently, Phytogbs were considered candidates for developing blood substitutes and as additives in veggie burgers.
References
- ↑ Kubo H., Uber hamoprotein aus den wurzelknollchen von leguminosen, Acta Phytochim. (Tokyo), 11 (1939) 195-200.
- ↑ Vainshtein B. K., Harutyunyan E. H., Kuranova I. P., Borisov V. V., N.I.Sosfenov, Pavlovsky A. G., Grebenko A. I.,Konareva N. V., Structure of leghaemoglobin from lupin root nodules at 5 A resolution., Nature, 254 (1975) 163-164
- 1 2 Hill R., Hargrove M. S.,Arredondo-Peter R., Phytoglobin: a novel nomenclature for plant globins accepted by the globin community at the 2014 XVIII conference on Oxygen-Binding and Sensing Proteins., F1000Research, 5 (2016) 212 (doi: 210.12688/f11000research.18133.12681).
- ↑ Martínez-Ocampo F., Vázquez-Limón C.,Arredondo-Peter R., Detection, PCR-amplification and characterization of a 4 intron-containing non-symbiotic hemoglobin gene from the moss Physcomitrella patens., Global J. Biochem., 3 (2012) 12
- ↑ Hardison R. C., A brief history of hemoglobins: plant, animal, protist, and bacteria., Proc. Natl. Acad. Sci. USA., 93 (1996) 5675-5679
- ↑ Go M., Correlation of DNA exonic regions with protein structural units in haemoglobin., Nature, 291 (1981) 90-92.
- ↑ Hyldig-Nielsen J. J., Jensen E., Paludan K., Wiborg O., Garret R., Joergersen P. L.,Marker K. A., The primary structures of two leghemoglobin genes from soybean, Nucleic Acids Res., 10 (1982) 689-701.
- ↑ Hargrove M., Brucker E. A., Stec B., Sarath G., Arredondo-Peter R., Klucas R. V., Olson J. S.,PhilipsJr. G. N., Crystal structure of a non-symbiotic hemoglobin., Structure, 8 (2000) 1005-1014.
- ↑ Reeder B. J.,Hough M. A., The structure of a class 3 nonsymbiotic plant haemoglobin from Arabidopsis thaliana reveals a novel N-terminal helical extension., Acta Crystallogr., D70 (2014) 1411-1418.
- ↑ Arredondo-Peter R., Hargrove M. S., Moran J. F., Sarath G.,Klucas R. V., Plant hemoglobins, Plant Physiol., 118 (1998) 1121-1126.
- ↑ Arredondo-Peter R., Hargrove M. S., Sarath G., Moran J. F., Lohrman J., Olson J. S.,Klucas R. V., Rice hemoglobins: gene cloning, analysis and oxygen-binding kinetics of a recombinant protein synthesized in Escherichia coli., Plant Physiol., 115 (1997) 1259-1266
- ↑ Wittenberg J. B., Bergersen F. J., Appleby C. A.,Turner G. L., Facilitated oxygen diffusion: the role of leghemoglobin in nitrogen fixation by bacteroids isolated from soybean root nodules., J. Biol. Chem., 249 (1974) 4057-4066
- ↑ Appleby C. A., Leghemoglobin and Rhizobium respiration., Annu. Rev. Plant Physiol., 35 (1984) 443-478
- ↑ Smagghe B. J., Trent J. T.,Hargrove M. S., NO dioxygenase activity in hemoglobins is ubiquitous in vitro, but limited by reduction in vivo., PLoS One, 3 (2008) doi: 10.1371/journal.pone.0002039.
- ↑ Hill R. D., Non-symbiotic haemoglobins-What´s happening beyond nitric oxide scavenging?, AoB Plants, Pls004 (2012) doi: 10.1093/aobpla/pls1004
- ↑ Garrocho-Villegas V.,Arredondo-Peter R., Molecular cloning and characterization of a moss (Ceratodon purpureus) non-symbiotic hemoglobin provides insight into the early evolution of plant non-symbiotic hemoglobins., Mol. Biol. Evol., 25 (2008) 1482-1487.
- ↑ Ross E. J. H., Shearman L., Mathiesen M., Zhou J., Arredondo-Peter R., Sarath G.,Klucas R. V., Non-symbiotic hemoglobins are synthesized during germination and in differentiating cell types., Protoplasma, 218 (2001) 125-133.
- ↑ Watts R. A., Hunt P. W., Hvitved A. N., Hargrove M. S., Peacock W. J.,Dennis E. S., A hemoglobin from plants homologous to truncated hemoglobins of microorganisms., Proc. Natl. Acad. Sci. USA., 98 (2001) 10119-10124.
- ↑ Taylor E. R., Nie X. Z., MacGregor A. W.,Hill R. D., A cereal haemoglobin gene is expressed in seed and root tissues under anaerobic conditions, Plant Mol. Biol., 24 (1994) 853-862
- ↑ Lira-Ruan V., Sarath G., Klucas R. V.,Arredondo-Peter R., Synthesis of hemoglobins in rice (Oryza sativa var. Jackson) plants growing in normal and stress conditions., Plant Sci., 161 (2001) 279-287.
- ↑ Vázquez-Limón C., Hoogewijs D., Vinogradov S. N.,Arredondo-Peter R., The evolution of land plant hemoglobins, Plant Sci., 191-192 (2012) 71-81.
- ↑ Vinogradov S. N., Fernández I., Hoogewijs D.,Arredondo-Peter R., Phylogenetic relationships of 3/3 and 2/2 hemoglobins in Archaeplastida genomes to bacterial and other eukaryote hemoglobins, Mol. Plant, 4 (2011) 42-58.
- ↑ Vinogradov S. N., Hoogewijs D.,Arredondo-Peter R., What are the origins and phylogeny of plant hemoglobins?, Commun. Integrat. Biol., 4 (2011) 443-445.