petite mutation
petite (ρ–) is a mutant first discovered in the yeast Saccharomyces cerevisiae. Due to the defect in the respiratory chain, 'petite' yeast are unable to grow on media containing only non-fermentable carbon sources (such as glycerol or ethanol) and form small colonies when grown in the presence of fermentable carbon sources (such as glucose). The petite phenotype can be caused by the absence of, or mutations in, mitochondrial DNA (termed "cytoplasmic Petites"), or by mutations in nuclear-encoded genes involved in oxidative phosphorylation[1]. A neutral petite produces all wild type progeny when crossed with wild type.
petite mutations can be induced using a variety of mutagens, including DNA intercalating agents, as well as chemicals that can interfere with DNA synthesis in growing cells. Mutagens that create Petites are implicated in increased rates of degenerative diseases and in the aging process.
Overview
A mutation that produces small (petite" > petite) anaerobic-like colonies had shown first in Yeast Saccharomyces cerevisiae and described by Boris Ephrussi and his co-workers in (1949a) in Gif-sur-Yvette, France.[2] The cells of petite colonies were smaller than those of wild-type colonies, but the term “petite” refers to colony not cell size.[2]
Causes
The petite caused by deficiency in cytochromes (a, a3 + b) and a lack of respiratory enzymes which engage in respiration in mitochondria.[3] Due to the error in the respiratory chain pathway, 'petite' yeast is incapable of growing on media containing only non-fermentable carbon sources (such as glycerol or ethanol) and forming small colonies when grown in the presence of fermentable carbon sources (such as glucose).[4] The absence of mitochondria can cause the petite phenotype, or mutations in mitochondrial DNA (termed "cytoplasmic Petites") which is a deletion mutation, or by mutations in nuclear-encoded genes involved in oxidative phosphorylation.
Experiment
Petite mutants can be generated in the laboratory by using high-efficiency treatments which are acriflavine, ethidium bromide, and other treatments.[5] Their mechanism is to break down and eventual loss of mitochondrial DNA: if the treatment time increases, the amount of mitochondrial DNA will decrease. After prolonged treatment, petites containing no detectable mitochondrial DNA were obtained.[3] It is useful approach to illustrate the function of mitochondrial DNA in yeast growth.
Petite mutation inheritance
The inheritance pattern of genes existing in the cell organelles such as mitochondria which named cytoplasmic inheritance differs from nuclear genes pattern.
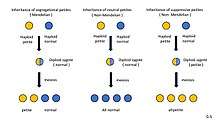
Petite mutation inheritance types
Petite mutants show extranuclear inheritance.The inheritance pattern varing with the type of petite involved.
Segregational petites (pet–): mutants are created by nuclear mutations and exhibit Mendelian 1:1 segregation.[5]
Neutral petites (rho–N): Neutral petite when crossed to wild-type, all offspring are wild-type. It has inherited normal mitochondrial DNA from wild-type parent, which is replicated in the offspring.[2]
Suppressive petites (rho–S): crosses between petite and wild-type, all offspring are petite, showing "dominant" behavior to suppress wild-type mitochondrial function.[2]
Most petite mutants are a suppressive type, and they differ from natural petite by affecting the wild-type, although both are a mutation in mitochondrial DNA. Mitochondrial genome of yeast will be the first eukaryotic genome to be understood in terms of both structure and function and this should smooth the way to understand the evolution of organelle genomes and its relationship with nuclear genomes.It is evident that Ephrussi’s work not only opened the field of extrachromosomal genetics, but also provide a fantastic incentive for the investigations which followed up to this day.[2]
References
- ↑ Lipinski, Kamil A.; Kaniak-Golik, Aneta; Golik, Pawel (June 2010). "Maintenance and expression of the S. cerevisiae mitochondrial genome—From genetics to evolution and systems biology". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1797 (6–7): 1086–1098. doi:10.1016/j.bbabio.2009.12.019. ISSN 0005-2728.
- 1 2 3 4 5 Bernardi, Giorgio (1979-09-01). "The petite mutation in yeast". Trends in Biochemical Sciences. 4 (9): 197–201. doi:10.1016/0968-0004(79)90079-3.
- 1 2 Goldring, Elizabeth S.; Grossman, Lawrence I.; Marmur, Julius (July 1971). "Petite Mutation in Yeast". Journal of Bacteriology. 107 (1): 377–381. ISSN 0021-9193. PMC 246929. PMID 5563875.
- ↑ Heslot, H.; Goffeau, A.; Louis, C. (1970-10-01). "Respiratory Metabolism of a "Petite Negative" Yeast Schizosaccharomyces pombe 972h−". Journal of Bacteriology. 104 (1): 473–481. ISSN 0021-9193. PMC 248232. PMID 4394400.
- 1 2 Heslot, H.; Louis, C.; Goffeau, A. (1970-10-01). "Segregational Respiratory-Deficient Mutants of a "Petite Negative" Yeast Schizosaccharomyces pombe 972h−". Journal of Bacteriology. 104 (1): 482–491. ISSN 0021-9193. PMC 248233. PMID 5473904.
- Ferguson, L.R.; von Borstel, R. C. (1992). "Induction of the cytoplasmic 'petite' mutation by chemical and physical agents in Saccharomyces cerevisiae". Mutation Research. 265 (1): 103–48. doi:10.1016/0027-5107(92)90042-z. PMID 1370239.