MUS81
Crossover junction endonuclease MUS81 is an enzyme that in humans is encoded by the MUS81 gene.[5][6][7]
In mammalian somatic cells, MUS81 and another structure specific DNA endonuclease, XPF (ERCC4), play overlapping and essential roles in completion of homologous recombination.[8] The significant overlap in function between these enzymes is most likely related to processing joint molecules such as D-loops and nicked Holliday junctions.[8]
Meiosis
MUS81 is a component of a minor chromosomal crossover (CO) pathway in the meiosis of budding yeast, plants and vertebrates.[9] However, in the protozoan Tetrahymena thermophila, MUS81 appears to be part of an essential (if not the predominant) CO pathway.[9] The MUS81 pathway also appears to be the predominant CO pathway in the fission yeast Schizosaccharomyces pombe.[9]
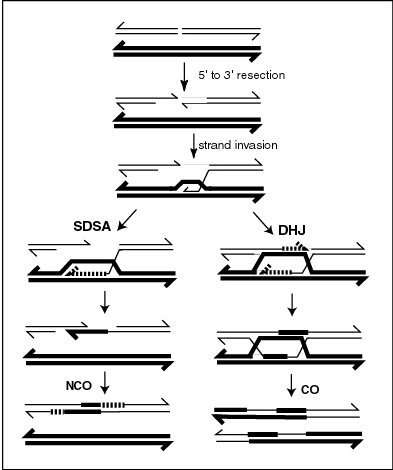
The relationship of the CO pathway to the overall process of meiotic recombination is illustrated in the accompanying diagram. Recombination during meiosis is often initiated by a DNA double-strand break (DSB). During recombination, sections of DNA at the 5' ends of the break are cut away in a process called resection. In the strand invasion step that follows, an overhanging 3' end of the broken DNA molecule "invades" the DNA of an homologous chromosome that is not broken forming a displacement loop (D-loop). After strand invasion, the further sequence of events may follow either of two main pathways, leading to a crossover (CO) or a non-crossover (NCO) recombinant (see Genetic recombination). The pathway leading to a CO involves a double Holliday junction (DHJ) intermediate. Holliday junctions need to be resolved for CO recombination to be completed.
MU81-MMS4, in the budding yeast Saccharomyces cerevisiae, is a DNA structure-selective endonuclease that cleaves joint DNA molecules formed during homologous recombination in meiosis and mitosis.[10] The MUS81-MMS4 endonuclease, although a minor resolvase for CO formation in S. cerevisiae, is crucial for limiting chromosome entanglements by suppressing multiple consecutive recombination events from initiating from the same DSB.[11]
Mus81 deficient mice have significant meiotic defects including the failure to repair a subset of DSBs.[12]
Interactions
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000172732 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000024906 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- 1 2 Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH (Nov 2001). "Human Mus81-associated endonuclease cleaves Holliday junctions in vitro". Molecular Cell. 8 (5): 1117–27. doi:10.1016/S1097-2765(01)00375-6. PMID 11741546.
- ↑ Constantinou A, Chen XB, McGowan CH, West SC (Oct 2002). "Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities". The EMBO Journal. 21 (20): 5577–85. doi:10.1093/emboj/cdf554. PMC 129086. PMID 12374758.
- ↑ "Entrez Gene: MUS81 MUS81 endonuclease homolog (S. cerevisiae)".
- 1 2 Kikuchi K, Narita T, Pham VT, Iijima J, Hirota K, Keka IS, Mohiuddin, Okawa K, Hori T, Fukagawa T, Essers J, Kanaar R, Whitby MC, Sugasawa K, Taniguchi Y, Kitagawa K, Takeda S (2013). "Structure-specific endonucleases xpf and mus81 play overlapping but essential roles in DNA repair by homologous recombination". Cancer Res. 73 (14): 4362–71. doi:10.1158/0008-5472.CAN-12-3154. PMC 3718858. PMID 23576554.
- 1 2 3 Lukaszewicz A, Howard-Till RA, Loidl J (2013). "Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex". Nucleic Acids Res. 41 (20): 9296–309. doi:10.1093/nar/gkt703. PMC 3814389. PMID 23935123.
- ↑ Mukherjee S, Wright WD, Ehmsen KT, Heyer WD (2014). "The Mus81-Mms4 structure-selective endonuclease requires nicked DNA junctions to undergo conformational changes and bend its DNA substrates for cleavage". Nucleic Acids Res. 42 (10): 6511–22. doi:10.1093/nar/gku265. PMC 4041439. PMID 24744239.
- ↑ Oke A, Anderson CM, Yam P, Fung JC (2014). "Controlling meiotic recombinational repair - specifying the roles of ZMMs, Sgs1 and Mus81/Mms4 in crossover formation". PLoS Genet. 10 (10): e1004690. doi:10.1371/journal.pgen.1004690. PMC 4199502. PMID 25329811.
- ↑ Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE (2008). "MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis". PLoS Genet. 4 (9): e1000186. doi:10.1371/journal.pgen.1000186. PMC 2525838. PMID 18787696.
Further reading
- Maruyama K, Sugano S (Jan 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (Oct 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P (Dec 2000). "Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1". Molecular and Cellular Biology. 20 (23): 8758–66. doi:10.1128/MCB.20.23.8758-8766.2000. PMC 86503. PMID 11073977.
- Oğrünç M, Sancar A (Jun 2003). "Identification and characterization of human MUS81-MMS4 structure-specific endonuclease". The Journal of Biological Chemistry. 278 (24): 21715–20. doi:10.1074/jbc.M302484200. PMID 12686547.
- Ciccia A, Constantinou A, West SC (Jul 2003). "Identification and characterization of the human mus81-eme1 endonuclease". The Journal of Biological Chemistry. 278 (27): 25172–8. doi:10.1074/jbc.M302882200. PMID 12721304.
- Blais V, Gao H, Elwell CA, Boddy MN, Gaillard PH, Russell P, McGowan CH (Feb 2004). "RNA interference inhibition of Mus81 reduces mitotic recombination in human cells". Molecular Biology of the Cell. 15 (2): 552–62. doi:10.1091/mbc.E03-08-0580. PMC 329235. PMID 14617801.
- Gao H, Chen XB, McGowan CH (Dec 2003). "Mus81 endonuclease localizes to nucleoli and to regions of DNA damage in human S-phase cells". Molecular Biology of the Cell. 14 (12): 4826–34. doi:10.1091/mbc.E03-05-0276. PMC 284787. PMID 14638871.
- Zhang R, Sengupta S, Yang Q, Linke SP, Yanaihara N, Bradsher J, Blais V, McGowan CH, Harris CC (Apr 2005). "BLM helicase facilitates Mus81 endonuclease activity in human cells". Cancer Research. 65 (7): 2526–31. doi:10.1158/0008-5472.CAN-04-2421. PMID 15805243.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T, Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai K, Isogai T, Sugano S (Jan 2006). "Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes". Genome Research. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- Hiyama T, Katsura M, Yoshihara T, Ishida M, Kinomura A, Tonda T, Asahara T, Miyagawa K (2006). "Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells". Nucleic Acids Research. 34 (3): 880–92. doi:10.1093/nar/gkj495. PMC 1360746. PMID 16456034.
- Ii M, Ii T, Brill SJ (Dec 2007). "Mus81 functions in the quality control of replication forks at the rDNA and is involved in the maintenance of rDNA repeat number in Saccharomyces cerevisiae". Mutation Research. 625 (1–2): 1–19. doi:10.1016/j.mrfmmm.2007.04.007. PMC 2100401. PMID 17555773.
- Nomura Y, Adachi N, Koyama H (Oct 2007). "Human Mus81 and FANCB independently contribute to repair of DNA damage during replication". Genes to Cells. 12 (10): 1111–22. doi:10.1111/j.1365-2443.2007.01124.x. PMID 17903171.