KIF2C
Kinesin-like protein KIF2C is a protein that in humans is encoded by the KIF2C gene.[5][6]
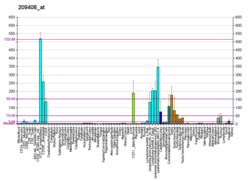
The protein encoded by this gene is a member of kinesin-like protein family. Most proteins of this family are microtubule-dependent molecular motors that transport organelles within cells and move chromosomes during cell division. This protein acts to regulate microtubule dynamics in cells and is important for anaphase chromosome segregation and may be required to coordinate the onset of sister centromere separation.[6]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000142945 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000028678 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Kim IG, Jun DY, Sohn U, Kim YH (Feb 1998). "Cloning and expression of human mitotic centromere-associated kinesin gene". Biochim Biophys Acta. 1359 (3): 181–6. doi:10.1016/s0167-4889(97)00103-1. PMID 9434124.
- 1 2 "Entrez Gene: KIF2C kinesin family member 2C".
Further reading
- Miki H, Setou M, Kaneshiro K, Hirokawa N (2001). "All kinesin superfamily protein, KIF, genes in mouse and human". Proc. Natl. Acad. Sci. U.S.A. 98 (13): 7004–11. doi:10.1073/pnas.111145398. PMC 34614. PMID 11416179.
- Maney T, Hunter AW, Wagenbach M, Wordeman L (1998). "Mitotic Centromere–associated Kinesin Is Important for Anaphase Chromosome Segregation". J. Cell Biol. 142 (3): 787–801. doi:10.1083/jcb.142.3.787. PMC 2148171. PMID 9700166.
- Wordeman L, Wagenbach M, Maney T (2000). "Mutations in the ATP-binding domain affect the subcellular distribution of mitotic centromere-associated kinesin (MCAK)". Cell Biol. Int. 23 (4): 275–86. doi:10.1006/cbir.1999.0359. PMID 10600236.
- Sugata N, Li S, Earnshaw WC, et al. (2001). "Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere--kinetochore complexes". Hum. Mol. Genet. 9 (19): 2919–26. doi:10.1093/hmg/9.19.2919. PMID 11092768.
- Maney T, Wagenbach M, Wordeman L (2001). "Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin". J. Biol. Chem. 276 (37): 34753–8. doi:10.1074/jbc.M106626200. PMID 11466324.
- Cheng LJ, Zhou ZM, Li JM, et al. (2002). "Expression of a novel HsMCAK mRNA splice variant, tsMCAK gene, in human testis". Life Sci. 71 (23): 2741–57. doi:10.1016/S0024-3205(02)02079-9. PMID 12383881.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Cassimeris L, Morabito J (2004). "TOGp, the Human Homolog of XMAP215/Dis1, Is Required for Centrosome Integrity, Spindle Pole Organization, and Bipolar Spindle Assembly". Mol. Biol. Cell. 15 (4): 1580–90. doi:10.1091/mbc.E03-07-0544. PMC 379257. PMID 14718566.
- Ganem NJ, Compton DA (2004). "The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK". J. Cell Biol. 166 (4): 473–8. doi:10.1083/jcb.200404012. PMC 2172212. PMID 15302853.
- Newton CN, Wagenbach M, Ovechkina Y, et al. (2004). "MCAK, a Kin I kinesin, increases the catastrophe frequency of steady-state HeLa cell microtubules in an ATP-dependent manner in vitro". FEBS Lett. 572 (1–3): 80–4. doi:10.1016/j.febslet.2004.06.093. PMID 15304328.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Andersen JS, Lam YW, Leung AK, et al. (2005). "Nucleolar proteome dynamics". Nature. 433 (7021): 77–83. doi:10.1038/nature03207. PMID 15635413.
- Moore AT, Rankin KE, von Dassow G, et al. (2005). "MCAK associates with the tips of polymerizing microtubules". J. Cell Biol. 169 (3): 391–7. doi:10.1083/jcb.200411089. PMC 2171944. PMID 15883193.
- Rual JF, Venkatesan K, Hao T, et al. (2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Ganem NJ, Upton K, Compton DA (2006). "Efficient mitosis in human cells lacking poleward microtubule flux". Curr. Biol. 15 (20): 1827–32. doi:10.1016/j.cub.2005.08.065. PMID 16243029.
- Helenius J, Brouhard G, Kalaidzidis Y, et al. (2006). "The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends". Nature. 441 (7089): 115–9. doi:10.1038/nature04736. PMID 16672973.
- Sun Y, Huang YC, Xu QZ, et al. (2006). "HIV-1 Tat depresses DNA-PK(CS) expression and DNA repair, and sensitizes cells to ionizing radiation". Int. J. Radiat. Oncol. Biol. Phys. 65 (3): 842–50. doi:10.1016/j.ijrobp.2006.02.040. PMID 16751065.
- Olsen JV, Blagoev B, Gnad F, et al. (2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.