Brome mosaic virus
| Brome mosaic virus | |
|---|---|
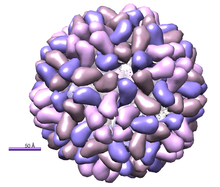 | |
| Crystal structure of Brome Mosaic Virus. PDB entry 1js9[1] | |
| Virus classification | |
| Group: | Group IV ((+)ssRNA) |
| Family: | Bromoviridae |
| Genus: | Bromovirus |
| Species: | Brome mosaic virus |
Brome mosaic virus (BMV) is a small (28 nm, 86S), positive-stranded, icosahedral RNA plant virus belonging to the genus Bromovirus, family Bromoviridae, in the alphavirus-like superfamily.
BMV commonly infects Bromus inermis (see Bromus) and other grasses, can be found almost anywhere wheat is grown, and thrives in areas with heavy foot or machinery traffic. It is also one of the few grass viruses that infects dicotyledonous plants, such as soybean [2]; however, it primarily infects monocotyledonous plants, such as barley and others in the family Gramineae.
BMV was first isolated in 1942 from bromegrass (Bromus inermis),[3] had its genomic organization determined by the 1970s, and was completely sequenced with commercially available clones by the 1980s.[4][5]
The alphavirus-like superfamily includes more than 250 plant and animal viruses including Tobacco mosaic virus, Semliki forest virus, Hepatitis E virus, Sindbis virus, and arboviruses (which cause certain types of encephalitis).[6][7] Many of the positive-strand RNA viruses that belong to the alphavirus family share a high degree of similarity in proteins involved in genomic replication and synthesis.[8][9] The sequence similarities of RNA replication genes and strategies for BMV have been shown to extend to a wide range of plant and animal viruses beyond the alphaviruses, including many other positive-strand RNA viruses from other families.[10] Understanding how these viruses replicate and targeting key points in their life cycle can help advance antiviral treatments worldwide.
BMV has a genome that is divided into three 5' capped RNAs. RNA1 (3.2 kb) encodes a protein called 1a (109 kDa), which contains both an N-proximal methyltransferase domain and a C-proximal helicase-like domain. The methyltransferase domain shows sequence similarity to other alphavirus m7G methyltransferases and guanyltransferases, called nsP1 proteins, involved in RNA capping.[11] RNA2 (2.9 kb) encodes the 2a protein (94 kDa), the RNA-dependent RNA polymerase, responsible for replication of the viral genome.[10][6] The dicistronic RNA3 (2.1 kb) encodes for two proteins, the 3a protein (involved in cell-to-cell migration during infection) and the coat protein (for RNA encapsidation and vascular spread), which is expressed from a subgenomic replication intermediate mRNA, called RNA4 (0.9 kb). 3a and coat protein are essential for systemic infection in plants but not RNA replication.[12][6][13]
References
- ↑ Lucas, R. W.; Larson, S. B.; McPherson, A. (2002). "The crystallographic structure of brome mosaic virus". Journal of Molecular Biology. 317 (1): 95–108. doi:10.1006/jmbi.2001.5389. PMID 11916381.
- ↑ Díaz-Cruz, G.A.; Smith, C.M.; Wiebe, K.F.; Charette, J.M.; Cassone, B.J. (2018). "First Report of Brome mosaic virus Infecting Soybean, Isolated in Manitoba, Canada". Plant Disease. 102 (2): 460. doi:10.1094/PDIS-07-17-1012-PDN.
- ↑ Lane, L. C. (1974). "The bromoviruses". Advances in virus research. 19: 151–220. doi:10.1016/s0065-3527(08)60660-0. PMID 4613160.
- ↑ Ahlquist, P.; Luckow, V.; Kaesberg, P. (1981). "Complete nucleotide sequence of brome mosaic virus RNA3". Journal of Molecular Biology. 153 (1): 23–38. doi:10.1016/0022-2836(81)90524-6. PMID 7338913.
- ↑ Lane, 2003
- 1 2 3 Sullivan & Ahlquist, 1997
- ↑ Lampio, 1999
- ↑ Ahlquist, P.; Strauss, E. G.; Rice, C. M.; Strauss, J. H.; Haseloff, J.; Zimmern, D. (1985). "Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses". Journal of Virology. 53 (2): 536–542. PMC 254668. PMID 3968720.
- ↑ French, R.; Ahlquist, P. (1988). "Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA". Journal of Virology. 62 (7): 2411–2420. PMC 253399. PMID 3373573.
- 1 2 Ahlquist, P. (1992). "Bromovirus RNA replication and transcription". Current Opinion in Genetics & Development. 2 (1): 71–76. doi:10.1016/S0959-437X(05)80325-9. PMID 1378769.
- ↑ Ahola, T.; Ahlquist, P. (1999). "Putative RNA capping activities encoded by brome mosaic virus: Methylation and covalent binding of guanylate by replicase protein 1a". Journal of Virology. 73 (12): 10061–10069. PMC 113057. PMID 10559320.
- ↑ Sacher, R.; Ahlquist, P. (1989). "Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection". Journal of Virology. 63 (11): 4545–4552. PMC 251087. PMID 2795712.
- ↑ Diez, J.; Ishikawa, M.; Kaido, M.; Ahlquist, P. (2000). "Identification and characterization of a host protein required for efficient template selection in viral RNA replication". Proceedings of the National Academy of Sciences. 97 (8): 3913–3918. doi:10.1073/pnas.080072997. PMC 18116. PMID 10759565.
- Charles J. Sailey. Portion of thesis entitled Characterization of Brome Mosaic Virus RNA3 interaction with GCD10, a tRNA binding host factor from yeast. 2005. University of the Sciences in Philadelphia.