ALDH1A1
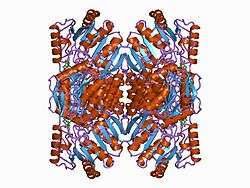
Aldehyde dehydrogenase 1 family, member A1, also known as ALDH1A1 or retinaldehyde dehydrogenase 1 (RALDH1), is an enzyme that in humans is encoded by the ALDH1A1 gene.[5][6]
Function
This protein belongs to the aldehyde dehydrogenases family of proteins. Aldehyde dehydrogenase is the second enzyme of the major oxidative pathway of alcohol metabolism. Two major liver isoforms of this enzyme, cytosolic and mitochondrial, can be distinguished by their electrophoretic mobilities, kinetic properties, and subcellular localizations. Most Caucasians have two major isozymes, while approximately 50% of East Asians have only the cytosolic isozyme, missing the mitochondrial isozyme. A remarkably higher frequency of acute alcohol intoxication among East Asians than among Caucasians could be related to the absence of the mitochondrial isozyme. This gene encodes the main cytosolic isoform, which has a lower affinity for aldehydes than the mitochondrial enzyme.[7]
ALDH1A1 also belongs to the group of corneal crystallins that help maintain the transparency of the cornea.[8]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000165092 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000053279 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Pereira F, Rosenmann E, Nylen E, Kaufman M, Pinsky L, Wrogemann K (March 1991). "The 56 kDa androgen binding protein is an aldehyde dehydrogenase". Biochem. Biophys. Res. Commun. 175 (3): 831–8. doi:10.1016/0006-291X(91)91640-X. PMID 1709013.
- ↑ Hsu LC, Tani K, Fujiyoshi T, Kurachi K, Yoshida A (June 1985). "Cloning of cDNAs for human aldehyde dehydrogenases 1 and 2". Proc. Natl. Acad. Sci. U.S.A. 82 (11): 3771–5. doi:10.1073/pnas.82.11.3771. PMC 397869. PMID 2987944.
- ↑ "Entrez Gene: ALDH1A1".
- ↑ Jester JV, Moller-Pedersen T, Huang J, Sax CM, Kays WT, Cavangh HD, Petroll WM, Piatigorsky J (March 1999). "The cellular basis of corneal transparency: evidence for 'corneal crystallins'". J. Cell Sci. 112. ( Pt 5): 613–22. PMID 9973596.
External links
- Human ALDH1A1 genome location and ALDH1A1 gene details page in the UCSC Genome Browser.
Further reading
- Walsh N, Dowling P, O'Donovan N, et al. (2008). "Aldehyde dehydrogenase 1A1 and gelsolin identified as novel invasion-modulating factors in conditioned medium of pancreatic cancer cells". Journal of proteomics. 71 (5): 561–71. doi:10.1016/j.jprot.2008.09.002. PMID 18848913.
- Barley K, Dracheva S, Byne W (2009). "Subcortical oligodendrocyte- and astrocyte-associated gene expression in subjects with schizophrenia, major depression and bipolar disorder". Schizophr. Res. 112 (1–3): 54–64. doi:10.1016/j.schres.2009.04.019. PMID 19447584.
- Ekhart C, Doodeman VD, Rodenhuis S, et al. (2008). "Influence of polymorphisms of drug metabolizing enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the pharmacokinetics of cyclophosphamide and 4-hydroxycyclophosphamide". Pharmacogenet. Genomics. 18 (6): 515–23. doi:10.1097/FPC.0b013e3282fc9766. PMID 18496131.
- Rodriguez FJ, Giannini C, Asmann YW, et al. (2008). "Gene expression profiling of NF-1-associated and sporadic pilocytic astrocytoma identifies aldehyde dehydrogenase 1 family member L1 (ALDH1L1) as an underexpressed candidate biomarker in aggressive subtypes". J. Neuropathol. Exp. Neurol. 67 (12): 1194–204. doi:10.1097/NEN.0b013e31818fbe1e. PMC 2730602. PMID 19018242.
- Carpentino JE, Hynes MJ, Appelman HD, et al. (2009). "Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer". Cancer Res. 69 (20): 8208–15. doi:10.1158/0008-5472.CAN-09-1132. PMC 2776663. PMID 19808966.
- Ma S, Chan KW, Lee TK, et al. (2008). "Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations". Mol. Cancer Res. 6 (7): 1146–53. doi:10.1158/1541-7786.MCR-08-0035. PMID 18644979.
- Xiao T, Shoeb M, Siddiqui MS, et al. (2009). "Molecular cloning and oxidative modification of human lens ALDH1A1: implication in impaired detoxification of lipid aldehydes". J. Toxicol. Environ. Health Part A. 72 (9): 577–84. doi:10.1080/15287390802706371. PMID 19296407.
- Cañestro C, Catchen JM, Rodríguez-Marí A, et al. (2009). Gojobori T, ed. "Consequences of lineage-specific gene loss on functional evolution of surviving paralogs: ALDH1A and retinoic acid signaling in vertebrate genomes". PLoS Genet. 5 (5): e1000496. doi:10.1371/journal.pgen.1000496. PMC 2682703. PMID 19478994.
- Saito A, Kawamoto M, Kamatani N (2009). "Association study between single-nucleotide polymorphisms in 199 drug-related genes and commonly measured quantitative traits of 752 healthy Japanese subjects". J. Hum. Genet. 54 (6): 317–23. doi:10.1038/jhg.2009.31. PMID 19343046.
- Chen YC, Chen YW, Hsu HS, et al. (2009). "Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer". Biochem. Biophys. Res. Commun. 385 (3): 307–13. doi:10.1016/j.bbrc.2009.05.048. PMID 19450560.
- Tabakoff B, Saba L, Printz M, et al. (2009). "Genetical genomic determinants of alcohol consumption in rats and humans". BMC Biol. 7: 70. doi:10.1186/1741-7007-7-70. PMC 2777866. PMID 19874574.
- Morimoto K, Kim SJ, Tanei T, et al. (2009). "Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression". Cancer Sci. 100 (6): 1062–8. doi:10.1111/j.1349-7006.2009.01151.x. PMID 19385968.
- Ekhart C, Rodenhuis S, Smits PH, et al. (2008). "Relations between polymorphisms in drug-metabolising enzymes and toxicity of chemotherapy with cyclophosphamide, thiotepa and carboplatin". Pharmacogenet. Genomics. 18 (11): 1009–15. doi:10.1097/FPC.0b013e328313aaa4. PMID 18854779.
- Wan C, Shi Y, Zhao X, et al. (2009). "Positive association between ALDH1A2 and schizophrenia in the Chinese population". Prog. Neuropsychopharmacol. Biol. Psychiatry. 33 (8): 1491–5. doi:10.1016/j.pnpbp.2009.08.008. PMID 19703508.
- Low SK, Kiyotani K, Mushiroda T, et al. (2009). "Association study of genetic polymorphism in ABCC4 with cyclophosphamide-induced adverse drug reactions in breast cancer patients". J. Hum. Genet. 54 (10): 564–71. doi:10.1038/jhg.2009.79. PMID 19696793.
- Moore SM, Liang T, Graves TJ, et al. (2009). "Identification of a novel cytosolic aldehyde dehydrogenase allele, ALDH1A1*4". Hum. Genomics. 3 (4): 304–7. doi:10.1186/1479-7364-3-4-304. PMC 2885287. PMID 19706361.
- Chang B, Liu G, Xue F, et al. (2009). "ALDH1 expression correlates with favorable prognosis in ovarian cancers". Mod. Pathol. 22 (6): 817–23. doi:10.1038/modpathol.2009.35. PMC 2692456. PMID 19329942.
- Lind PA, Eriksson CJ, Wilhelmsen KC (2008). "The role of aldehyde dehydrogenase-1 (ALDH1A1) polymorphisms in harmful alcohol consumption in a Finnish population". Hum. Genomics. 3 (1): 24–35. doi:10.1186/1479-7364-3-1-24. PMID 19129088.
- Moreb JS, Baker HV, Chang LJ, et al. (2008). "ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells". Mol. Cancer. 7 (1): 87. doi:10.1186/1476-4598-7-87. PMC 2605459. PMID 19025616.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.