Chromista
Chromista is a biological kingdom consisting of some single-celled and multicellular eukaryotic organisms, which share similar features in their photosynthetic organelles (plastids).[1] It includes all protists such as some algae, diatoms, oomycetes, and protozoans whose plastids contain chlorophyll c. It is probably a polyphyletic group whose members independently arose as separate evolutionary group from the common ancestor of all eukaryotes.[2] As it is assumed the last common ancestor already possessed chloroplasts of red algal origin, the non-photosynthetic forms evolved from ancestors able to perform photosynthesis. Their plastids are surrounded by four membranes, and are believed to have been acquired from some red algae.
| Chromista | |
|---|---|
| Scientific classification | |
| Domain: | Eukaryota |
| (unranked): | |
| Kingdom: | Chromista Cavalier-Smith 1981 |
| Phyla | |
Chromista as a biological kingdom was created by British biologist Thomas Cavalier-Smith in 1981 to differentiate some protists from typical protozoans and plants.[3] According to Cavalier-Smith, the kingdom originally included only algae, but his later analysis indicated that many protozoans also belong to the new group. As of 2018, the kingdom is as diverse as kingdoms Plantae and Animalia, consisting of eight phyla. Notable members include marine algae, potato blight, dinoflagellates, Paramecium, brain parasite (Toxoplasma) and malarial parasite (Plasmodium).[4]
Biology
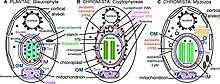
Members of Chromista are single-celled and multicellular eukaryotes having basically either or both features:[3]
- plastid(s) that contain chlorophyll c and lies within an extra (periplastid) membrane in the lumen of the rough endoplasmic reticulum (typically within the perinuclear cisterna);
- cilia with tripartite or bipartite rigid tubular hairs.
Even though the kingdom includes diverse organisms from algae to malarial parasites (Plasmodium), they are genetically related and are believed to have evolved from a common ancestor with all other eukaryotes but in an independent line of evolution. As a result of evolution, many have retained their plastids and cilia, while some have lost them.[5] Molecular evidences indicate that the plastids in chromists were derived from red algae through secondary symbiogenesis in a single event.[6] (In contrasts, plants acquired their plastids from cyanobacteria through primary symbiogenesis.[7]) These plastids are now enclosed in two extra cell membranes, making a four membrane envelope, as a result of which they acquired many other membrane proteins for transporting molecules in and out of the organelles. The diversity of chromists arose from degeneration, loss or replacement of the plastids in some lineages.[8] There was also additional symbiogenesis of green algae, the genes of which are retained in some members (such as heterokonts),[9] as well as bacterial chlorophyll (indicated by the presence of ribosomal protein L36 gene, rpl36) in haptophytes and cryptophytes.[10]
History and groups
The name Chromista was first introduced by Cavalier-Smith in 1981;[3] the earlier names Chromophyta, Chromobiota and Chromobionta correspond to roughly the same group.
It has been described as consisting of three different groups:[11]
- Heterokonts or stramenopiles: brown algae, diatoms, water moulds, etc.
- Haptophytes
- Cryptomonads
Cavalier-Smith later (in 2009) stated his reason for making a new kingdom, saying:
I established Chromista as a kingdom distinct from Plantae and Protozoa because of the evidence that chromist chloroplasts were acquired secondarily by enslavement of a red alga, itself a member of kingdom Plantae, and their unique membrane topology.[5]
Since then Chromista has been defined in different ways at different times. In 2010, Cavalier-Smith himself indicated his desire to move Alveolata, Rhizaria and Heliozoa into Chromista.
Some examples of classification of the Chromista and related groups are shown below.[12][13]
Chromophycées (Chadefaud, 1950)
The Chromophycées (Chadefaud, 1950),[14] renamed Chromophycota (Chadefaud, 1960),[15] included the current Ochrophyta (autotrophic Stramenopiles), Haptophyta (included in Chrysophyceae until Christensen, 1962), Cryptophyta, Dinophyta, Euglenophyceae and Choanoflagellida (included in Chrysophyceae until Hibberd, 1975).
Chromophyta (Christensen 1962, 1989)
The Chromophyta (Christensen 1962, 1989), defined as algae with chlorophyll c, included the current Ochrophyta (autotrophic Stramenopiles), Haptophyta, Cryptophyta, Dinophyta and Choanoflagellida. The Euglenophyceae were transferred to the Chlorophyta.
Chromophyta (Bourrelly, 1968)
The Chromophyta (Bourrelly, 1968) included the current Ochrophyta (autotrophic Stramenopiles), Haptophyta and Choanoflagellida. The Cryptophyceae and the Dinophyceae were part of Pyrrhophyta (= Dinophyta).
Chromista (Cavalier-Smith, 1981)
The Chromista (Cavalier-Smith, 1981) included the current Stramenopiles, Haptophyta and Cryptophyta.
Chromalveolata (Adl et al., 2005)
The Chromalveolata (Cavalier-Smith, 1981) included Stramenopiles, Haptophyta, Cryptophyta and Alveolata.
Classification
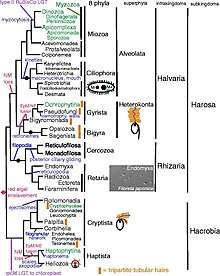
In his original classification in 1981, Cavalier-Smith included three phyla under Chromista, such as:[3]
- Heterokonta
- Haptophyta
- Cryptomonada
In 2010, Cavalier-Smith reorganised Chromista to include SAR supergroup such as (Stramenopiles, Alveolata and Rhizaria) and Hacrobia (Haptista, Cryptista).[5]
In 2017, he and his colleagues made a new higher-level grouping of all organisms as a revision of seven kingdoms model. In it, they classified Chromista into 2 subkingdoms and 10 phyla, namely:[16]
- Subkingdom Hacrobia
- Phulym N.N. (=nomen nominandum, name unknown)
- Phylum Cryptista
- Phylum Haptophyta
- Phylum Heliozoa
- Subkingdom Harosa (=Supergroup SAR)
- Phylum Ciliophora
- Phylum Miozoa
- Phylum Bigyra
- Phylum Pseudofungi (=Oomycota)
- Phylum Cercozoa
- Phylum Retaria
Cavalier-Smith made a new analysis of Chromista in 2018 in which he classified all chromists into 8 phyla, as follows:[4]
- Phylum Miozoa
- Phylum Ciliophora
- Phylum Gyrista
- Phylum Bigyra
- Phylum Cercozoa
- Phylum Retaria
- Phylum Cryptista
- Phylum Haptista
Phylogeny
| Chromista |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Controversy
Molecular trees have had some difficulty resolving relationships between the different groups. All three may share a common ancestor with the alveolates (see chromalveolates), but there is evidence that suggests that the haptophytes and cryptomonads do not belong together with the heterokonts or the SAR clade, but may be associated with the Archaeplastida.[1][17] Cryptista specifically may be sister or part of Archaeplastida.[18]
References
- Parfrey LW, Barbero E, Lasser E, Dunthorn M, Bhattacharya D, Patterson DJ, Katz LA (December 2006). "Evaluating support for the current classification of eukaryotic diversity". PLOS Genetics. 2 (12): e220. doi:10.1371/journal.pgen.0020220. PMC 1713255. PMID 17194223.
- Cavalier-Smith T, Allsopp MT, Chao EE (November 1994). "Chimeric conundra: are nucleomorphs and chromists monophyletic or polyphyletic?". Proceedings of the National Academy of Sciences of the United States of America. 91 (24): 11368–72. Bibcode:1994PNAS...9111368C. doi:10.1073/pnas.91.24.11368. PMC 45232. PMID 7972066.
- Cavalier-Smith T (1981). "Eukaryote kingdoms: seven or nine?". Bio Systems. 14 (3–4): 461–81. doi:10.1016/0303-2647(81)90050-2. PMID 7337818.
- Cavalier-Smith, Thomas (2018). "Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences". Protoplasma. 255 (1): 297–357. doi:10.1007/s00709-017-1147-3. PMC 5756292. PMID 28875267.
- Cavalier-Smith, Thomas (2009). "Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree". Biology Letters. 6 (3): 342–345. doi:10.1098/rsbl.2009.0948. PMC 2880060. PMID 20031978.
- Keeling, Patrick J. (2009). "Chromalveolates and the Evolution of Plastids by Secondary Endosymbiosis". Journal of Eukaryotic Microbiology. 56 (1): 1–8. doi:10.1111/j.1550-7408.2008.00371.x. PMID 19335769.
- Ponce-Toledo, Rafael I.; Deschamps, Philippe; López-García, Purificación; Zivanovic, Yvan; Benzerara, Karim; Moreira, David (2017). "An Early-Branching Freshwater Cyanobacterium at the Origin of Plastids". Current Biology. 27 (3): 386–391. doi:10.1016/j.cub.2016.11.056. PMC 5650054. PMID 28132810.
- Keeling, Patrick J. (2010). "The endosymbiotic origin, diversification and fate of plastids". Philosophical Transactions of the Royal Society B: Biological Sciences. 365 (1541): 729–748. doi:10.1098/rstb.2009.0103. PMC 2817223. PMID 20124341.
- Morozov, A.A.; Galachyants, Yuri P. (2019). "Diatom genes originating from red and green algae: Implications for the secondary endosymbiosis models". Marine Genomics. 45: 72–78. doi:10.1016/j.margen.2019.02.003. PMID 30792089.
- Rice, Danny W; Palmer, Jeffrey D (2006). "An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters". BMC Biology. 4 (1): 31. doi:10.1186/1741-7007-4-31. PMC 1570145. PMID 16956407.
- Csurös M, Rogozin IB, Koonin EV (May 2008). "Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach". Molecular Biology and Evolution. 25 (5): 903–11. doi:10.1093/molbev/msn039. PMID 18296415.
- de Reviers B (2006). Biologia e Filogenia das Algas. Porto Alegre: Editora Artmed. pp. 156–157. ISBN 9788536315102.
- Blackwell W (2009). "Chromista revisited: a dilemma of overlapping putative kingdoms, and the attempted application of the botanical code of nomenclature" (PDF). Phytologia. 91 (2): 191–225.
- Chadefaud M (1950). "Les cellules nageuses des Algues dans l'embranchement des Chromophycées" [Seaweed swimming cells in the branch of Chromophyceae]. Comptes rendus hebdomadaires des séances de l'Académie des Sciences (in French). 231: 788–790.
- Chadefaud M (1960). "Les végétaux non vasculaires (Cryptogamie)". In Chadefaud M, Emberger L (eds.). Traité de Botanique Systématique. Paris: Tome I.
- Ruggiero, Michael A.; Gordon, Dennis P.; Orrell, Thomas M.; Bailly, Nicolas; Bourgoin, Thierry; Brusca, Richard C.; Cavalier-Smith, Thomas; Guiry, Michael D.; Kirk, Paul M.; Thuesen, Erik V. (2015). "A higher level classification of all living organisms". PLOS ONE. 10 (4): e0119248. Bibcode:2015PLoSO..1019248R. doi:10.1371/journal.pone.0119248. PMC 4418965. PMID 25923521.
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J (August 2007). "Phylogenomics reshuffles the eukaryotic supergroups". PLOS ONE. 2 (8): e790. Bibcode:2007PLoSO...2..790B. doi:10.1371/journal.pone.0000790. PMC 1949142. PMID 17726520.
- Burki F, Kaplan M, Tikhonenkov DV, Zlatogursky V, Minh BQ, Radaykina LV, Smirnov A, Mylnikov AP, Keeling PJ (January 2016). "Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista". Proceedings. Biological Sciences. 283 (1823): 20152802. doi:10.1098/rspb.2015.2802. PMC 4795036. PMID 26817772.