Carbon monoxide dehydrogenase
In enzymology, carbon monoxide dehydrogenase (CODH) (EC 1.2.7.4) is an enzyme that catalyzes the chemical reaction
- CO + H2O + A CO2 + AH2
| carbon-monoxide dehydrogenase (acceptor) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.2.7.4 | ||||||||
| CAS number | 64972-88-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
The chemical process catalyzed by carbon monoxide dehydrogenase is referred to as a water-gas shift reaction.
The 3 substrates of this enzyme are CO, H2O, and A, whereas its two products are CO2 and AH2.
A variety of electron donors/receivers (Shown as "A" and "AH2" in the reaction equation above) are observed in micro-organisms which utilize CODH. Several examples of electron transfer cofactors include Ferredoxin, NADP+/NADPH and flavoprotein complexes like flavin adenine dinucleotide (FAD).[1][2][3] Carbon monoxide dehydrogenases support the metabolisms of diverse prokaryotes, including methanogens, aerobic carboxidotrophs, acetogens, sulfate-reducers, and hydrogenogenic bacteria. The bidirectional reaction catalyzed by CODH plays a role in the carbon cycle allowing organisms to both make use of CO as a source of energy and utilize CO2 as a source of carbon. CODH can form a monofunctional enzyme, as is the case in Rhodospirillum rubram, or can form a cluster with acetyl-CoA synthase as has been shown in M.thermoacetica. When acting in concert, either as structurally independent enzymes or in a bifunctional CODH/ACS unit, the two catalytic sites are key to carbon fixation in the reductive acetyl-CoA pathway Microbial organisms (Both Aerobic and Anaerobic) encode and synthesize CODH for the purpose of carbon fixation (CO oxidation and CO2 reduction). Depending on attached accessory proteins (A,B,C,D-Clusters), serve a variety of catalytic functions, including reduction of [4Fe-4S] clusters and insertion of Nnckel.[4]
This enzyme belongs to the family of oxidoreductases, specifically those acting on the aldehyde or oxo group of donor with other acceptors. The systematic name of this enzyme class is carbon-monoxide:acceptor oxidoreductase. Other names in common use include anaerobic carbon monoxide dehydrogenase, carbon monoxide oxygenase, carbon-monoxide dehydrogenase, and carbon-monoxide:(acceptor) oxidoreductase.
Classes
Aerobic carboxydotrophic bacteria utilize copper-molybdenum flavoenzymes. Anaerobic bacteria utilize nickel-iron based CODHs due to their oxygen sensitive natur. CODH containing a Mo-[2Fe-2S]-FAD active site have been found in aerobic bacteria, while a distinct class of Ni-[3Fe-4S] CODH enzymes have been purified from anaerobic bacteria.[5][6][7] Both classes of CODH catalyze the reversible conversion between carbon dioxide (CO2) and carbonmonoxide (CO). CODH exists in both monofuctional and bifunctional forms. In the latter case, CODH forms a bifunctional cluster with acetyl-CoA synthase, as has been well characterized in the anaerobic bacteria Moorella thermoacetica.[8][9]
Structure
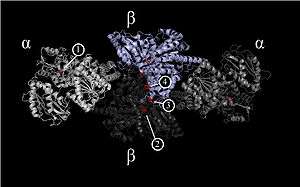
Homodimeric Ni-CODH consists of five metal complexes referred to as clusters. Each differing in individual coordination geometry, presence of nickel, and location of the active site in either sub-unit α or β.[10] Multiple research groups have proposed crystal structures for the α2β2 tetrameric enzyme CODH/ACS from the acetogenic bacteria M. thermoacetica, including two recent examples since 2009: 3I01 2Z8Y. The two β units are the site of CODH activity and form the central core of the enzyme. In total, the 310 kDa enzyme contains seven iron-sulfur [4Fe-4S] clusters. Each α unit contains a single metal cluster. Together, the two β units contains five clusters of three types. CODH catalytic activity occurs at the Ni-[3Fe-4S] C-clusters while the interior [4Fe-4S] B and D clusters transfer electrons away from the C-cluster to external electron carriers such as ferredoxin. The ACS activity occurs in A-cluster located in the outer two α units.[6][7]
A noteworthy feature of the M. thermoacetica CODH/ACS is an internal gas tunnel connecting the multiple active sites.[11] The full role of the gas channel in regulating the rate catalytic activity is still a subject of investigation, but several studies support the notion that molecules of CO do in fact travel directly from the C-cluster to the ACS active site without leaving the enzyme. For instance, the rate of acetyl-CoA synthase activity in the bifunctional enzyme is not affected by the addition of hemoglobin, which would compete for CO in bulk solution,[12] and isotopic labeling studies show that carbon monoxide derived from the C-cluster is preferentially used at the A-cluster over unlabeled CO in solution.[13] Protein engineering of the CODH/ACS in M.thermoacetica revealed that mutating residues, so as to functionally block the tunnel, stopped acety-CoA synthesis when only CO2 was present.[14] The discovery of a functional CO tunnel places CODH on a growing list of enzymes that independently evolved this strategy to transfer reactive intermediates from one active site to another.[15]
Reaction mechanisms
Oxidative
The CODH catalytic site, referred to as the C-cluster, is a [3Fe-4S] cluster bonded to a Ni-Fe moiety. Two basic amino acids (Lys587 and His 113 in M.thermoacetica) reside in proximity to the C-cluster and facilitate acid-base chemistry required for enzyme activity.[16] Based on IR spectra suggesting the presence of an Ni-CO complex, the proposed first step in the oxidative catalysis of CO to CO2 involves the binding of CO to Ni2+ and corresponding complexing of Fe2+ to a water molecule.[17]
The binding of CO molecule causes a shift in the coordination of the Ni atom from a square-planar to square pyramidal geometry.[18] Dobbek et al. further propose that movement of the nickel atom’s cysteine ligand brings the CO into close proximity to the hydroxyl group, and facilitate a base-catalyzed, nucleophillic attack by the iron-bound hydroxy group. A carboxy bridge between the Ni and Fe atom has been proposed as an intermediate.[19] A decarboxylation leads to the release of CO2 and the reduction of the cluster. Although the resulting intermediate oxidation state of the Ni and the degree to which electrons are distributed throughout the Ni-[3Fe-4S] cluster is subject of some debate, the electrons in the reduced C-cluster are transferred to nearby B and D [4Fe-4S] clusters, returning the Ni-[3Fe-4S] C-cluster to an oxidized state and reducing the single electron carrier ferredoxin.[20][21]
Reductive
Given CODH's role in CO2 fixation, it is common in the biochemistry literature for the reductive mechanism to be inferred as the “direct reverse” of the oxidative mechanism by the ”principal of microreversibility.”[22] In the process of reducing carbon dioxide, the enzyme's C-cluster must first be activated from an oxidized to a reduced state before the Ni-CO2 bond is formed.[23]
Environmental relevance
Carbon monoxide dehydrogenase is closely associated with the regulation of atmospheric CO and CO2 levels, maintaining optimal CO levels suitable for other forms of life. Microbial organisms rely on these enzymes for both energy conservation along with CO2 fixation. Often encoding for and synthesizing multiple unique forms of CODH for designated use. Further research into specific types of CODH show CO being used and condensed with CH3 (Methyl groups) to form Acetyl-CoA.[24] Anaerobic micro-organisms like Acetogens undergo the Wood-Ljungdahl Pathway, relying on CODH to produce CO by reduction of CO2 needed for the synthesis of Acetyl-CoA from a methyl, coenzyme a (CoA) and corrinoid iron-sulfur protein.[25] Other types show CODH being utilized to generate a proton motive force for the purposes of energy generation. CODH is used for the CO oxidation, producing two protons which are subsequently reduced to form dihydrogen (H2, known colloquially as molecular hydrogen), providing the necessary free energy to drive ATP generation.[26]
References
- Buckel W, Thauer RK (2018). "+ (Rnf) as Electron Acceptors: A Historical Review". Frontiers in Microbiology. 9: 401. doi:10.3389/fmicb.2018.00401. PMC 5861303. PMID 29593673.
- Kracke F, Virdis B, Bernhardt PV, Rabaey K, Krömer JO (December 2016). "Clostridium autoethanogenum by extracellular electron supply". Biotechnology for Biofuels. 9 (1): 249. doi:10.1186/s13068-016-0663-2. PMC 5112729. PMID 27882076.
- van den Berg WA, Hagen WR, van Dongen WM (February 2000). "The hybrid-cluster protein ('prismane protein') from Escherichia coli. Characterization of the hybrid-cluster protein, redox properties of the [2Fe-2S] and [4Fe-2S-2O] clusters and identification of an associated NADH oxidoreductase containing FAD and [2Fe-2S]". European Journal of Biochemistry. 267 (3): 666–76. doi:10.1046/j.1432-1327.2000.01032.x. PMID 10651802.
- Hadj-Saïd J, Pandelia ME, Léger C, Fourmond V, Dementin S (December 2015). "The Carbon Monoxide Dehydrogenase from Desulfovibrio vulgaris". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1847 (12): 1574–83. doi:10.1016/j.bbabio.2015.08.002. PMID 26255854.
- Jeoung J, Fesseler J, Goetzl S, Dobbek H (2014). "Chapter 3. Carbon Monoxide. Toxic Gas and Fuel for Anaerobes and Aerobes: Carbon Monoxide Dehydrogenases". In Kroneck PM, Torres ME (eds.). The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences. 14. Springer. pp. 37–69. doi:10.1007/978-94-017-9269-1_3. PMID 25416390.
- Dobbek H, Svetlitchnyi V, Gremer L, Huber R, Meyer O (August 2001). "Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster". Science. 293 (5533): 1281–5. Bibcode:2001Sci...293.1281D. doi:10.1126/science.1061500. PMID 11509720. S2CID 21633407.
- Ragsdale S (September 2010). Sigel H, Sigel A (ed.). "Metal-Carbon Bonds in Enzymes and Cofactors". Coordination Chemistry Reviews. Metal Ions in Life Sciences. Royal Society of Chemistry. 254 (17–18): 1948–1949. doi:10.1039/9781847559333. ISBN 978-1-84755-915-9. PMC 2923820. PMID 20729977.
- Doukov TI, Blasiak LC, Seravalli J, Ragsdale SW, Drennan CL (March 2008). "Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase". Biochemistry. 47 (11): 3474–83. doi:10.1021/bi702386t. PMC 3040099. PMID 18293927.
- Tan X, Volbeda A, Fontecilla-Camps JC, Lindahl PA (April 2006). "Function of the tunnel in acetylcoenzyme A synthase/carbon monoxide dehydrogenase". Journal of Biological Inorganic Chemistry. 11 (3): 371–8. doi:10.1007/s00775-006-0086-9. PMID 16502006.
- Wittenborn EC, Merrouch M, Ueda C, Fradale L, Léger C, Fourmond V, et al. (October 2018). "Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase". eLife. 7: e39451. doi:10.7554/eLife.39451. PMC 6168284. PMID 30277213.
- Doukov TI, Blasiak LC, Seravalli J, Ragsdale SW, Drennan CL (March 2008). "Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase". Biochemistry. 47 (11): 3474–83. doi:10.1021/bi702386t. PMC 3040099. PMID 18293927.
- Doukov TI, Iverson TM, Seravalli J, Ragsdale SW, Drennan CL (October 2002). "A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase" (PDF). Science. 298 (5593): 567–72. Bibcode:2002Sci...298..567D. doi:10.1126/science.1075843. PMID 12386327.
- Seravalli J, Ragsdale SW (February 2000). "Channeling of carbon monoxide during anaerobic carbon dioxide fixation". Biochemistry. 39 (6): 1274–7. doi:10.1021/bi991812e. PMID 10684606.
- Tan X, Loke HK, Fitch S, Lindahl PA (April 2005). "The tunnel of acetyl-coenzyme a synthase/carbon monoxide dehydrogenase regulates delivery of CO to the active site". Journal of the American Chemical Society. 127 (16): 5833–9. doi:10.1021/ja043701v. PMID 15839681.
- Weeks A, Lund L, Raushel FM (October 2006). "Tunneling of intermediates in enzyme-catalyzed reactions". Current Opinion in Chemical Biology. 10 (5): 465–72. doi:10.1016/j.cbpa.2006.08.008. PMID 16931112.
- Ragsdale SW (August 2006). "Metals and their scaffolds to promote difficult enzymatic reactions". Chemical Reviews. 106 (8): 3317–37. doi:10.1021/cr0503153. PMID 16895330.
- Chen J, Huang S, Seravalli J, Gutzman H, Swartz DJ, Ragsdale SW, Bagley KA (December 2003). "Infrared studies of carbon monoxide binding to carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica". Biochemistry. 42 (50): 14822–30. doi:10.1021/bi0349470. PMID 14674756.
- Dobbek H, Svetlitchnyi V, Gremer L, Huber R, Meyer O (August 2001). "Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster". Science. 293 (5533): 1281–5. Bibcode:2001Sci...293.1281D. doi:10.1126/science.1061500. PMID 11509720. S2CID 21633407.
- Ha SW, Korbas M, Klepsch M, Meyer-Klaucke W, Meyer O, Svetlitchnyi V (April 2007). "Interaction of potassium cyanide with the [Ni-4Fe-5S] active site cluster of CO dehydrogenase from Carboxydothermus hydrogenoformans". The Journal of Biological Chemistry. 282 (14): 10639–46. doi:10.1074/jbc.M610641200. PMID 17277357.
- Wang VC, Ragsdale SW, Armstrong FA (2014). "Investigations of the efficient electrocatalytic interconversions of carbon dioxide and carbon monoxide by nickel-containing carbon monoxide dehydrogenases". In Peter M.H. Kroneck, Martha E. Sosa Torres (eds.). The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences. 14. Springer. pp. 71–97. doi:10.1007/978-94-017-9269-1_4. ISBN 978-94-017-9268-4. PMC 4261625. PMID 25416391.
- Ragsdale SW (November 2007). "Nickel and the carbon cycle". Journal of Inorganic Biochemistry. 101 (11–12): 1657–66. doi:10.1016/j.jinorgbio.2007.07.014. PMC 2100024. PMID 17716738.
- Ragsdale SW, Pierce E (December 2008). "Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1784 (12): 1873–98. doi:10.1016/j.bbapap.2008.08.012. PMC 2646786. PMID 18801467.
- Feng J, Lindahl PA (February 2004). "Carbon monoxide dehydrogenase from Rhodospirillum rubrum: effect of redox potential on catalysis". Biochemistry. 43 (6): 1552–9. doi:10.1021/bi0357199. PMID 14769031.
- Hadj-Saïd J, Pandelia ME, Léger C, Fourmond V, Dementin S (December 2015). "The Carbon Monoxide Dehydrogenase from Desulfovibrio vulgaris". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1847 (12): 1574–83. doi:10.1016/j.bbabio.2015.08.002. PMID 26255854.
- Ragsdale SW, Pierce E (December 2008). "Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1784 (12): 1873–98. doi:10.1016/j.bbapap.2008.08.012. PMC 2646786. PMID 18801467.
- Ensign SA, Ludden PW (September 1991). "Characterization of the CO oxidation/H2 evolution system of Rhodospirillum rubrum. Role of a 22-kDa iron-sulfur protein in mediating electron transfer between carbon monoxide dehydrogenase and hydrogenase". The Journal of Biological Chemistry. 266 (27): 18395–403. PMID 1917963.
Further reading
- Drennan CL, Heo J, Sintchak MD, Schreiter E, Ludden PW (October 2001). "Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase". Proceedings of the National Academy of Sciences of the United States of America. 98 (21): 11973–8. Bibcode:2001PNAS...9811973D. doi:10.1073/pnas.211429998. PMC 59822. PMID 11593006.
- Jeoung JH, Martins BM, Dobbek H (2019). "Carbon Monoxide Dehydrogenases". In Hu Y (ed.). Metalloproteins. Methods in Molecular Biology. 1876. New York: Springer. pp. 37–54. doi:10.1007/978-1-4939-8864-8_3. ISBN 9781493988631. PMID 30317473.
- Jeoung JH, Dobbek H (November 2007). "Carbon dioxide activation at the Ni,Fe-cluster of anaerobic carbon monoxide dehydrogenase". Science. American Association for the Advancement of Science. 318 (5855): 1461–4. Bibcode:2007Sci...318.1461J. doi:10.1126/science.1148481. JSTOR 20051712. PMID 18048691. S2CID 41063549.
- Dobbek H, Svetlitchnyi V, Gremer L, Huber R, Meyer O (August 2001). "Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster". Science. 293 (5533): 1281–5. Bibcode:2001Sci...293.1281D. doi:10.1126/science.1061500. PMID 11509720. S2CID 21633407.* Hegg EL (October 2004). "Unraveling the structure and mechanism of acetyl-coenzyme A synthase". Accounts of Chemical Research. 37 (10): 775–83. doi:10.1021/ar040002e. PMID 15491124. S2CID 29401674.*
- Hu Z, Spangler NJ, Anderson ME, Xia J, Ludden PW, Lindahl PA, Münck E (1996-01-01). "Nature of the C-Cluster in Ni-Containing Carbon Monoxide Dehydrogenases". Journal of the American Chemical Society. 118 (4): 830–845. doi:10.1021/ja9528386. ISSN 0002-7863.
- Bartholomew GW, Alexander M (May 1979). "Microbial metabolism of carbon monoxide in culture and in soil". Applied and Environmental Microbiology. 37 (5): 932–7. doi:10.1128/AEM.37.5.932-937.1979. PMC 243327. PMID 485139.