Antibody-dependent enhancement
Antibody-dependent enhancement (ADE), sometimes less precisely called immune enhancement or disease enhancement, is a phenomenon in which binding of a virus to non-neutralizing antibodies enhances its entry into host cells, and sometimes also its replication.[1] This phenomenon—which leads to both increased infectivity and virulence—has been observed with mosquito-borne flaviviruses such as Dengue virus, Yellow fever virus and Zika virus,[2][3] with HIV, and with coronaviruses.[4]

There are various hypotheses on how ADE happens and there is a likelihood that more than one mechanism exists. In one such pathway, some cells of the immune system lack the usual receptors on their surfaces that the virus uses to gain entry, but they have Fc receptors that bind to one end of antibodies. The virus binds to the antigen-binding site at the other end, and in this way gains entry to and infects the immune cell. Dengue virus can use this mechanism to infect human macrophages, if there was a preceding infection with a different strain of the virus, causing a normally mild viral infection to become life-threatening.[5]
An ongoing question in the COVID-19 pandemic is whether—and if so, to what extent—COVID-19 receives ADE from prior infection with other coronaviruses.[6]
ADE can hamper vaccine development, as a vaccine may cause the production of antibodies which, via ADE, worsen the disease the vaccine is designed to protect against. Vaccine candidates for Dengue virus,[4] and feline infectious peritonitis virus (a cat coronavirus) had to be stopped because they elicited ADE.[7]
In coronavirus infection
Alphacoronavirus
The feline infectious peritonitis virus (FIPV) is an alphacoronavirus that is a very common pathogen in both domestic and wild cats.[8] It is believed that FIPV can cause antibody-dependent enhancement (ADE). Thus, vaccination against FIPV surprisingly increases the disease seriousness.[9] Moreover, it was demonstrated that infection of macrophages by FIPV in vitro can be triggered by non-neutralising monoclonal antibodies targeting the Spike (S)-protein, and this phenomenon can also occur with diluted neutralising antibodies.[10] ADE explains why half of cats develop peritonitis after being passively immunized with antivirus antibodies and being challenged with the same FIPV serotype.[11] In several countries an attenuated virus vaccine is available in a form of nasal drops; however, its application is still considered controversial by many experts, both in terms of safety and efficacy.[12] The mechanism of antibody-dependent enhancement for FIPV is illustrated in the video.
Betacoronavirus
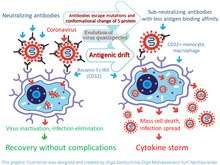

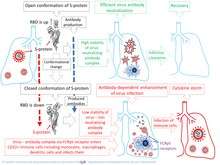
The neutralization ability of an antibody on a virion is dependent on concentration and the strength of interaction between antibody and antigen. High-affinity antibodies can cause virus neutralization by recognizing specific viral epitopes. However, pathogen-specific antibodies can promote a phenomenon known as antibody-dependent enhancement (ADE), which can be induced when the strength of antibody-antigen interaction is below the certain threshold.[14][15]
There are multiple examples of ADE triggered by betacoronaviruses.[14][15]
ADE related vaccine development problems
The development of immunopathology upon exposure has been a major challenge for coronavirus vaccine development[16] and may similarly impact SARS-CoV-2 vaccine research. Non-human primates vaccinated with modified vaccinia Ankara virus encoding full-length SARS-CoV spike glycoprotein and challenged with the SARS-CoV virus had lower viral loads but suffered from acute lung injury due to ADE.[17] ADE has been observed in both severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) animal models allowing the respective viruses to enter cells expressing Fc𝛾R including myeloid lineage cells.[18]
ADE of acute lung injury has been documented in animal models of both SARS and MERS. Rabbits intranasally infected with MERS-CoV developed a pulmonary infection characterized by viremia and perivascular inflammation of the lung, and an antibody response that lacked neutralizing antibodies.[19] The rabbits developed more severe lung disease on re-exposure to MERS-CoV, and developed neutralizing antibodies after reinfection.[19] In SARS, mice vaccinated with four types of vaccines against SARS-CoV, as well as those infected with SARS-CoV itself, developed neutralizing antibodies.[16] Mice were then challenged with live SARS-CoV, upon which all developed immunopathologic-type lung disease, although none had detectable virus two days after challenge and were protected compared to control.[16]
It was shown that antibodies triggered by a SARS-CoV vaccine candidate, represented by recombinant, full-length SARS-CoV Spike-protein trimers, promote infection of immune cell lines.[20] Fluorescence microscopy and real-time quantitative reverse transcriptase polymerase chain reaction (RT-PCR) were used for demonstration that anti-Spike immune serum increases infection of human monocyte-derived macrophages by SARS-CoV.[20] Antibody-mediated infection was dependent on binding of immune complexes to cell surface FcγRII. Intact cytoplasmic signaling domains FcγRII were important for sustaining ADE of SARS-CoV infection. The authors of the study[20] concluded that primary human immune cells where susceptible to infection by SARS-CoV in the presence of anti-Spike antibodies. The SARS-CoV-1 virus (SARS-CoV) can enter macrophages via an antibody-mediated pathway and can replicate in these cells.[21]
Mechanism of ADE for coronavirus infections
ADE in coronavirus infection can be caused by conformation changes of spike (S)-protein or/and high mutation rate of the gene that encodes spike (S) protein. A thorough analysis of amino acid variability in SARS-CoV-2 virus proteins, that included the S-protein, revealed that least conservative amino acids are in most exposed fragments of S-protein including receptor binding domain (RBD). Therefore, antigenic drift is a most likely cause of amino-acids variability in this protein [15][22] and ADE. This drift can occur in a form of changes of both types of antigenic epitopes, including conformational and linear.
Potential linkage between pathophysiology of COVID-19 and ADE
The pathophysiology of SARS and COVID-19 diseases may be associated with ADE. The authors of the study[15] believe that ADE is a key step in the progression of disease from its mild to severe form. Onset of ADE, due to antigenic drift, can explain the observed sudden immune dysregulation, including apoptosis of immune cells, which promotes the development of T-cell lymphopenia and an inflammatory cascade with the lung accumulation of macrophages and neutrophils, as well as a cytokine storm. ADE goes along with reduction of Th1 cytokines IL2, TNF-α and IFN-γ and increase of Th2 cytokines IL-10, IL-6, PGE-2 and INF-α, as well as with inhibition of STAT pathway.[23]
Perhaps ADE is the reason why the course of SARS and COVID-19 is more severe for older people compared to younger people. It is likely that in older people the production of antibodies is slower and by the time the antibodies are developed in the titer that is sufficient to neutralize the virus, the virus changes its antigenic determinants. In this case, immuno-dominant neutralizing antibodies might start forming unstable complexes with the new form of the virus and start to infect monocytes/macrophages causing ADE. This process can trigger generalized infection of immune cells in multiple organs and cytokine storm.[24][25]
It is interesting that in mice similar phenomenon of developing more severe disease in old compared to young animals exists. In contrast to old mice, in young mice, despite detectable viral replication оf SARS-CoV-1 in the lungs upon infection, clinical signs of the disease do not develop.[26]
In influenza infection
Prior receipt of 2008–09 TIV (Trivalent Inactivated Influenza Vaccine) was associated with an increased risk of medically attended pH1N1 illness during the spring-summer 2009 in Canada. The occurrence of bias (selection, information) or confounding cannot be ruled out. Further experimental and epidemiological assessment is warranted. Possible biological mechanisms and immunoepidemiologic implications are considered.[27]
Natural infection and the attenuated vaccine induce antibodies that enhance the update of the homologous virus and H1N1 virus isolated several years later, demonstrating that a primary influenza A virus infection results in the induction of infection enhancing antibodies.[28]
ADE was suspected in infections with influenza A virus subtype H7N9, but knowledge is limited.
In dengue virus infection
The most widely known example of ADE occurs in the setting of infection with dengue virus, a single-stranded positive-polarity RNA virus of the family Flaviviridae. It causes a disease of varying severity in humans, from dengue fever (DF), which is usually self-limited, to dengue hemorrhagic fever and dengue shock syndrome, either of which may be life-threatening.[29] It is estimated that as many as 390 million individuals are infected with dengue virus annually.[30]
The phenomenon of ADE may be observed when a person who has previously been infected with one serotype of the dengue virus becomes infected months or years later with a different serotype. In such cases, the clinical course of the disease is more severe, and these people have higher viremia compared with those in whom ADE has not occurred. This explains the observation that while primary (first) infections cause mostly minor disease (dengue fever) in children, secondary infection (re-infection at a later date) is more likely to be associated with dengue hemorrhagic fever and/or dengue shock syndrome in both children and adults.[31]
There are four antigenically different serotypes of dengue virus (dengue virus 1–4).[32] In 2013 a fifth serotype was reported.[33] Infection with dengue virus induces the production of neutralizing homotypic immunoglobulin G (IgG) antibodies which provide lifelong immunity against the infecting serotype. Infection with dengue virus also produces some degree of cross-protective immunity against the other three serotypes.[34] Neutralizing heterotypic (cross-reactive) IgG antibodies are responsible for this cross-protective immunity, which typically persists for a period of several months to a few years. These heterotypic antibody titers decrease over long time periods (4 to 20 years).[35] While heterotypic IgG antibody titers decrease, homotypic IgG antibody titers increase over long time periods. This could be due to the preferential survival of long-lived memory B cells producing homotypic antibodies.[35]
In addition to inducing neutralizing heterotypic antibodies, infection with the dengue virus can also induce heterotypic antibodies that neutralize the virus only partially or not at all.[36] The production of such cross-reactive but non-neutralizing antibodies could be the reason for more severe secondary infections. It is thought that by binding to but not neutralizing the virus, these antibodies cause it to behave as a "trojan horse",[37][38][39] where it is delivered into the wrong compartment of dendritic cells that have ingested the virus for destruction.[40][41] Once inside the white blood cell, the virus replicates undetected, eventually generating very high virus titers which cause severe disease.[42]
A study conducted by Modhiran et al.[43] attempted to explain how non-neutralizing antibodies down-regulate the immune response in the host cell through the Toll-like receptor signaling pathway. Toll-like receptors are known to recognize extra- and intracellular viral particles and to be a major basis of the cytokines production. In vitro experiments showed that the inflammatory cytokines and type 1 interferon production were reduced when the ADE-dengue virus complex bound to the Fc receptor of THP-1 cells. This can be explained by both a decrease of Toll-like receptor production and a modification of its signaling pathway. On one hand, an unknown protein induced by the stimulated Fc receptor reduces the Toll-like receptor transcription and translation, which reduces the capacity of the cell to detect viral proteins. On the other hand, many proteins (TRIF, TRAF6, TRAM, TIRAP, IKKα, TAB1, TAB2, NF-κB complex) involved in the Toll-like receptor signaling pathway are down-regulated, which led to a decrease of the cytokine production. Two of them, TRIF and TRAF6, are respectively down-regulated by 2 proteins SARM and TANK up-regulated by the stimulated Fc receptors.
To illustrate the phenomenon of ADE, consider the following example: an epidemic of dengue fever occurred in Cuba, lasting from 1977 to 1979. The infecting serotype was dengue virus-1. This epidemic was followed by two more outbreaks of dengue fever—one in 1981 and one in 1997; dengue virus-2 was the infecting serotype in both of these later epidemics. 205 cases of dengue hemorrhagic fever and dengue shock syndrome occurred during the 1997 outbreak, all in people older than 15 years. All but three of these cases were demonstrated to have been previously infected by the dengue virus-1 serotype during the epidemic of 1977–1979.[44] Furthermore, people who had been infected with dengue virus-1 during the 1977-79 outbreak and secondarily infected with dengue virus-2 in 1997 had a 3-4 fold increased probability of developing severe disease than those secondarily infected with dengue virus-2 in 1981.[35] This scenario can be explained by the presence of neutralizing heterotypic IgG antibodies in sufficient titers in 1981, the titers of which had decreased by 1997 to the point where they no longer provided significant cross-protective immunity.
In HIV-1 virus infection
ADE of infection has also been reported in HIV. Like dengue virus, non-neutralizing level of antibodies have been found to enhance the viral infection through interactions of the complement system and receptors.[45] The increase in infection has been reported to be over 350 fold which is comparable to ADE in other viruses like dengue virus.[45] ADE in HIV can be complement-mediated or Fc receptor-mediated. Complements in the presence of HIV-1 positive sera have been found to enhance the infection of MT-2 T-cell line. The Fc-receptor mediated enhancement was reported when HIV infection was enhanced by sera from HIV-1 positive guinea pig enhanced the infection of peripheral blood mononuclear cells without the presence of any complements.[46] Complement component receptors CR2, CR3 and CR4 have been found to mediate this Complement-mediated enhancement of infection.[45][47] The infection of HIV-1 leads to activation of complements. Fragments of these complements can assist viruses with infection by facilitating viral interactions with host cells that express complement receptors.[48] The deposition of complement on the virus brings the gp120 protein close to CD4 molecules on the surface of the cells, thus leading to facilitated viral entry.[48] Viruses pre-exposed to non-neutralizing complement system have also been found to enhance infections in interdigitating dendritic cells. Opsonized viruses have not only shown enhanced entry but also favorable signaling cascades for HIV replication in interdigitating dendritic cells.[49]
HIV-1 has also shown enhancement of infection in HT-29 cells when the viruses were pre-opsonized with complements C3 and C9 in seminal fluid. This enhanced rate of infection was almost 2 times greater than infection of HT-29 cells with virus alone.[50] Subramanian et al., reported that almost 72% of serum samples out of 39 HIV positive individuals contained complements that were known to enhance the infection. They also suggested that the presence of neutralizing antibody or antibody-dependent cellular cytotoxicity-mediating antibodies in the serum contains infection-enhancing antibodies.[51] The balance between the neutralizing antibodies and infection-enhancing antibodies changes as the disease progresses. During advanced stages of the disease the proportion of infection-enhancing antibodies are generally higher than neutralizing antibodies.[52] Increase in viral protein synthesis and RNA production have been reported to occur during the complement-mediated enhancement of infection. Cells that are challenged with non-neutralizing levels of complements have been found have accelerated release of reverse transcriptase and the viral progeny.[53] The interaction of anti-HIV antibodies with non-neutralizing complement exposed viruses also aid in binding of the virus and the erythrocytes which can lead to more efficient delivery of viruses to the immune-compromised organs.[47]
ADE in HIV has raised questions about the risk of infections to volunteers who have taken sub-neutralizing levels of vaccine just like any other viruses that exhibit ADE. Gilbert et al., in 2005 reported that there was no ADE of infection when they used rgp120 vaccine in phase 1 and 2 trials.[54] It has been emphasized that much research needs to be done in the field of the immune response to HIV-1, information from these studies can be used to produce a more effective vaccine.
Mechanism
There are several possibilities to explain the phenomenon:
- A viral surface protein studded with antibodies against a virus of one serotype binds to a similar virus with a different serotype. The binding is meant to neutralize the virus surface protein from attaching to the cell, but the virus-antibody complex also binds to the Fc-region antibody receptor (FcγR) on the cell membrane. This brings the virus into close proximity to the virus-specific receptor, and the cell internalizes the virus through the normal infection route.[55]
- A virus surface protein may be attached to antibodies of a different serotype, activating the classical pathway of the complement system. The complement cascade system instead binds C1Q complex attached to the virus surface protein via the antibodies, which in turn bind C1q receptor found on cells, bringing the virus and the cell close enough for a specific virus receptor to bind the virus, beginning infection. This mechanism has not been shown specifically for dengue virus infection, but may occur with Ebola virus infection in vitro.[56]
- When an antibody to a virus is present for a different serotype, it is unable to neutralize the virus, which is then ingested into the cell as a sub-neutralized virus particle. These viruses are phagocytosed as antigen-antibody complexes, and degraded by macrophages. Upon ingestion the antibodies no longer even sub-neutralize the body due to the denaturing condition at the step for acidification of phagosome before fusion with lysosome. The virus becomes active and begins its proliferation within the cell.
See also
- Original antigenic sin
- Other ways in which antibodies can (unusually) make an infection worse instead of better
- Blocking antibody, which can be either good or bad, depending on circumstances
- Hook effect, most relevant to in vitro tests but known to have some in vivo relevances
References
- Tirado, S. M.; Yoon, K. J. (2003). "Antibody-dependent enhancement of virus infection and disease". Viral Immunology. 16 (1): 69–86. doi:10.1089/088282403763635465. PMID 12725690.
- Khandia, R.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Malik, Y. S.; Singh, R. K.; Chaicumpa, W. (2018). "Modulation of Dengue/Zika Virus Pathogenicity by Antibody-Dependent Enhancement and Strategies to Protect Against Enhancement in Zika Virus Infection". Frontiers in Immunology. 9: 597. doi:10.3389/fimmu.2018.00597. PMC 5925603. PMID 29740424.
- Plotkin, S; Orenstein, W (2012). "Yellow fever vaccine". Vaccines (6 ed.). Amsterdam: Elsevier. pp. 870–968. ISBN 9781455700905.
- Ahuja, Anjana (17 June 2020). "The long road to a Covid-19 vaccine". Financial Times. Retrieved 17 June 2020.
- Dimmock NJ, Easton AJ, Leppard K (2007). Introduction to modern virology. Malden, MA: Blackwell Pub. p. 65. ISBN 978-1-4051-3645-7.
- Tetro JA (March 2020). "Is COVID-19 receiving ADE from other coronaviruses?". Microbes and Infection. 22 (2): 72–73. doi:10.1016/j.micinf.2020.02.006. PMC 7102551. PMID 32092539.
- Peeples, Lynne (14 April 2020). "News Feature: Avoiding pitfalls in the pursuit of a COVID-19 vaccine". Proceedings of the National Academy of Sciences. 117 (15): 8218–8221. doi:10.1073/pnas.2005456117. ISSN 0027-8424. PMC 7165470. PMID 32229574.
- Vennema, Harry; Poland, Amy; Foley, Janet; Pedersen, Niels C. (March 1998). "Feline Infectious Peritonitis Viruses Arise by Mutation from Endemic Feline Enteric Coronaviruses". Virology. 243 (1): 150–157. doi:10.1006/viro.1998.9045. ISSN 0042-6822. PMC 7131759. PMID 9527924.
- Vennema, H; de Groot, R J; Harbour, D A; Dalderup, M; Gruffydd-Jones, T; Horzinek, M C; Spaan, W J (1990). "Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization". Journal of Virology. 64 (3): 1407–1409. doi:10.1128/jvi.64.3.1407-1409.1990. ISSN 0022-538X.
- Hohdatsu, T.; Nakamura, M.; Ishizuka, Y.; Yamada, H.; Koyama, H. (September 1991). "A study on the mechanism of antibody-dependent enhancement of feline infectious peritonitis virus infection in feline macrophages by monoclonal antibodies". Archives of Virology. 120 (3–4): 207–217. doi:10.1007/bf01310476. ISSN 0304-8608. PMC 7087175. PMID 1659798.
- TAKANO, Tomomi; YAMADA, Shinji; DOKI, Tomoyoshi; HOHDATSU, Tsutomu (2019). "Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: Antibody-dependent enhancement infection of cats with type I FIPV via the oral route". Journal of Veterinary Medical Science. 81 (6): 911–915. doi:10.1292/jvms.18-0702. ISSN 0916-7250. PMID 31019150.
- Negro, Francesco (2020-04-16). "Is antibody-dependent enhancement playing a role in COVID-19 pathogenesis?". Swiss Medical Weekly. doi:10.4414/smw.2020.20249. ISSN 1424-3997. PMID 32298458.
- Ricke, Darrell; Malone, Robert W. (2020). "Medical Countermeasures Analysis of 2019-nCoV and Vaccine Risks for Antibody-Dependent Enhancement (ADE)". SSRN Working Paper Series. doi:10.2139/ssrn.3546070. ISSN 1556-5068.
- Iwasaki, Akiko; Yang, Yexin (2020-04-21). "The potential danger of suboptimal antibody responses in COVID-19". Nature Reviews Immunology. doi:10.1038/s41577-020-0321-6. ISSN 1474-1733. PMID 32317716.
- Ricke, Darrell; Malone, Robert W. (2020). "Medical Countermeasures Analysis of 2019-nCoV and Vaccine Risks for Antibody-Dependent Enhancement (ADE)". SSRN Working Paper Series. doi:10.2139/ssrn.3546070. ISSN 1556-5068.
- Tseng C, et al. (2012). "Immunization with SARS Coronavirus Vaccines Leads to Pulmonary Immunopathology on Challenge with the SARS Virus". PLOS ONE. 7 (4): e35421. Bibcode:2012PLoSO...735421T. doi:10.1371/journal.pone.0035421. PMC 3335060. PMID 22536382.
- Liu L, et al. (2019). "Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection". JCI Insight. 4 (4). doi:10.1172/jci.insight.123158. PMC 6478436. PMID 30830861.
- Wan Y, et al. (2020). "Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry". Journal of Virology. 94 (5). doi:10.1128/JVI.02015-19. PMC 7022351. PMID 31826992.
- Houser KV, et al. (2017). "Enhanced inflammation in New Zealand white rabbits when MERS-CoV reinfection occurs in the absence of neutralizing antibody". PLOS Pathogens. 13 (8): e1006565. doi:10.1371/journal.ppat.1006565. PMC 5574614. PMID 28817732.
- Leung, Hiu-lan, Nancy. Mechanism of antibody-dependent enhancement in severe acute respiratory syndrome coronavirus infection (Thesis). The University of Hong Kong Libraries. doi:10.5353/th_b4732706.
- Yip, Ming S; Cheung, Chung Y; Li, Ping H; Bruzzone, Roberto; Peiris, JS Malik; Jaume, Martial (2011-01-10). "Investigation of Antibody-Dependent Enhancement (ADE) of SARS coronavirus infection and its role in pathogenesis of SARS". BMC Proceedings. 5 (S1). doi:10.1186/1753-6561-5-s1-p80. ISSN 1753-6561.
- Korber, B; Fischer, WM; Gnanakaran, S; Yoon, H; Theiler, J; Abfalterer, W; Foley, B; Giorgi, EE; Bhattacharya, T (2020-04-30). "Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2". doi:10.1101/2020.04.29.069054. Cite journal requires
|journal=(help) - Smatti, Maria K.; Al Thani, Asmaa A.; Yassine, Hadi M. (2018-12-05). "Viral-Induced Enhanced Disease Illness". Frontiers in Microbiology. 9: 2991. doi:10.3389/fmicb.2018.02991. ISSN 1664-302X. PMC 6290032. PMID 30568643.
- Gu, Jiang; Taylor, Clive R. (December 2003). "Acute Immunodeficiency, Multiple Organ Injury, and the Pathogenesis of SARS". Applied Immunohistochemistry & Molecular Morphology: 281–282. doi:10.1097/00129039-200312000-00001. ISSN 1541-2016. PMID 14663354.
- Gu, Jiang; Gong, Encong; Zhang, Bo; Zheng, Jie; Gao, Zifen; Zhong, Yanfeng; Zou, Wanzhong; Zhan, Jun; Wang, Shenglan; Xie, Zhigang; Zhuang, Hui (2005-08-01). "Multiple organ infection and the pathogenesis of SARS". The Journal of Experimental Medicine. 202 (3): 415–424. doi:10.1084/jem.20050828. ISSN 0022-1007. PMC 2213088. PMID 16043521.
- Sutton, Troy C.; Subbarao, Kanta (May 2015). "Development of animal models against emerging coronaviruses: From SARS to MERS coronavirus". Virology. 479-480: 247–258. doi:10.1016/j.virol.2015.02.030. ISSN 0042-6822. PMC 4793273. PMID 25791336.
- Skowronski DM, De Serres G, Crowcroft NS, Janjua NZ, Boulianne N, Hottes TS, et al. (April 2010). "Association between the 2008-09 seasonal influenza vaccine and pandemic H1N1 illness during Spring-Summer 2009: four observational studies from Canada". PLOS Medicine. 7 (4): e1000258. doi:10.1371/journal.pmed.1000258. PMC 2850386. PMID 20386731.
- Gotoff R, Tamura M, Janus J, Thompson J, Wright P, Ennis FA (January 1994). "Primary influenza A virus infection induces cross-reactive antibodies that enhance uptake of virus into Fc receptor-bearing cells". The Journal of Infectious Diseases. 169 (1): 200–3. doi:10.1093/infdis/169.1.200. PMID 8277183.
- Boonnak K, Slike BM, Burgess TH, Mason RM, Wu SJ, Sun P, et al. (April 2008). "Role of dendritic cells in antibody-dependent enhancement of dengue virus infection". Journal of Virology. 82 (8): 3939–51. doi:10.1128/JVI.02484-07. PMC 2292981. PMID 18272578.
- Ambuel Y, Young G, Brewoo JN, Paykel J, Weisgrau KL, Rakasz EG, et al. (15 September 2014). "A rapid immunization strategy with a live-attenuated tetravalent dengue vaccine elicits protective neutralizing antibody responses in non-human primates". Frontiers in Immunology. 5 (2014): 263. doi:10.3389/fimmu.2014.00263. PMC 4046319. PMID 24926294.
- Guzman MG, Vazquez S (December 2010). "The complexity of antibody-dependent enhancement of dengue virus infection". Viruses. 2 (12): 2649–62. doi:10.3390/v2122649. PMC 3185591. PMID 21994635.
- King CA, Anderson R, Marshall JS (August 2002). "Dengue virus selectively induces human mast cell chemokine production". Journal of Virology. 76 (16): 8408–19. doi:10.1128/JVI.76.16.8408-8419.2002. PMC 155122. PMID 12134044.
- Normile D (2013). "Tropical medicine. Surprising new Dengue virus throws a spanner in disease control efforts". Science. 342 (6157): 415. Bibcode:2013Sci...342..415N. doi:10.1126/science.342.6157.415. PMID 24159024.
- Alvarez G, Piñeros JG, Tobón A, Ríos A, Maestre A, Blair S, Carmona-Fonseca J (October 2006). "Efficacy of three chloroquine-primaquine regimens for treatment of Plasmodium vivax malaria in Colombia". The American Journal of Tropical Medicine and Hygiene. 75 (4): 605–9. doi:10.4269/ajtmh.2006.75.605. PMID 17038680.
- Guzman MG, Alvarez M, Rodriguez-Roche R, Bernardo L, Montes T, Vazquez S, et al. (February 2007). "Neutralizing antibodies after infection with dengue 1 virus". Emerging Infectious Diseases. 13 (2): 282–6. doi:10.3201/eid1302.060539. PMC 2725871. PMID 17479892.
- Goncalvez AP, Engle RE, St Claire M, Purcell RH, Lai CJ (May 2007). "Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention". Proceedings of the National Academy of Sciences of the United States of America. 104 (22): 9422–7. Bibcode:2007PNAS..104.9422G. doi:10.1073/pnas.0703498104. PMC 1868655. PMID 17517625.
- Peluso R, Haase A, Stowring L, Edwards M, Ventura P (November 1985). "A Trojan Horse mechanism for the spread of visna virus in monocytes". Virology. 147 (1): 231–6. doi:10.1016/0042-6822(85)90246-6. PMID 2998068.
- Chen YC, Wang SY (October 2002). "Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide". Journal of Virology. 76 (19): 9877–87. doi:10.1128/JVI.76.19.9877-9887.2002. PMC 136495. PMID 12208965.
- Witayathawornwong P (January 2005). "Fatal dengue encephalitis" (PDF). The Southeast Asian Journal of Tropical Medicine and Public Health. 36 (1): 200–2. PMID 15906668. Archived from the original (PDF) on 24 July 2011.
- Rodenhuis-Zybert IA, Wilschut J, Smit JM (August 2010). "Dengue virus life cycle: viral and host factors modulating infectivity". Cellular and Molecular Life Sciences. 67 (16): 2773–86. doi:10.1007/s00018-010-0357-z. PMID 20372965.
- Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, et al. (December 2010). "Dengue: a continuing global threat". Nature Reviews. Microbiology. 8 (12 Suppl): S7-16. doi:10.1038/nrmicro2460. PMC 4333201. PMID 21079655.
- Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, et al. (May 2010). "Cross-reacting antibodies enhance dengue virus infection in humans". Science. 328 (5979): 745–8. Bibcode:2010Sci...328..745D. doi:10.1126/science.1185181. PMC 3837288. PMID 20448183.
- Modhiran N, Kalayanarooj S, Ubol S (December 2010). "Subversion of innate defenses by the interplay between DENV and pre-existing enhancing antibodies: TLRs signaling collapse". PLOS Neglected Tropical Diseases. PLOS ONE. 4 (12): e924. doi:10.1371/journal.pntd.0000924. PMC 3006139. PMID 21200427.
- Guzman MG (2000). "Dr. Guzman et al. Respond to Dr. Vaughn". American Journal of Epidemiology. 152 (9): 804. doi:10.1093/aje/152.9.804.
- Willey S, Aasa-Chapman MM, O'Farrell S, Pellegrino P, Williams I, Weiss RA, Neil SJ (March 2011). "Extensive complement-dependent enhancement of HIV-1 by autologous non-neutralising antibodies at early stages of infection". Retrovirology. 8: 16. doi:10.1186/1742-4690-8-16. PMC 3065417. PMID 21401915.
- Levy JA (2007). HIV and the pathogenesis of AIDS. Wiley-Blackwell. p. 247. ISBN 978-1-55581-393-2.
- Yu Q, Yu R, Qin X (September 2010). "The good and evil of complement activation in HIV-1 infection". Cellular & Molecular Immunology. 7 (5): 334–40. doi:10.1038/cmi.2010.8. PMC 4002684. PMID 20228834.
- Gras GS, Dormont D (January 1991). "Antibody-dependent and antibody-independent complement-mediated enhancement of human immunodeficiency virus type 1 infection in a human, Epstein-Barr virus-transformed B-lymphocytic cell line". Journal of Virology. 65 (1): 541–5. doi:10.1128/JVI.65.1.541-545.1991. PMC 240554. PMID 1845908.
- Bouhlal H, Chomont N, Réquena M, Nasreddine N, Saidi H, Legoff J, et al. (January 2007). "Opsonization of HIV with complement enhances infection of dendritic cells and viral transfer to CD4 T cells in a CR3 and DC-SIGN-dependent manner". Journal of Immunology. 178 (2): 1086–95. doi:10.4049/jimmunol.178.2.1086. PMID 17202372.
- Bouhlal H, Chomont N, Haeffner-Cavaillon N, Kazatchkine MD, Belec L, Hocini H (September 2002). "Opsonization of HIV-1 by semen complement enhances infection of human epithelial cells". Journal of Immunology. 169 (6): 3301–6. doi:10.4049/jimmunol.169.6.3301. PMID 12218150.
- Subbramanian RA, Xu J, Toma E, Morisset R, Cohen EA, Menezes J, Ahmad A (June 2002). "Comparison of human immunodeficiency virus (HIV)-specific infection-enhancing and -inhibiting antibodies in AIDS patients". Journal of Clinical Microbiology. 40 (6): 2141–6. doi:10.1128/JCM.40.6.2141-2146.2002. PMC 130693. PMID 12037078.
- Beck Z, Prohászka Z, Füst G (June 2008). "Traitors of the immune system-enhancing antibodies in HIV infection: their possible implication in HIV vaccine development". Vaccine. 26 (24): 3078–85. doi:10.1016/j.vaccine.2007.12.028. PMC 7115406. PMID 18241961.
- Robinson WE, Montefiori DC, Mitchell WM (April 1990). "Complement-mediated antibody-dependent enhancement of HIV-1 infection requires CD4 and complement receptors". Virology. 175 (2): 600–4. doi:10.1016/0042-6822(90)90449-2. PMID 2327077.
- Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, et al. (March 2005). "Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial". The Journal of Infectious Diseases. 191 (5): 666–77. doi:10.1086/428405. PMID 15688279.
- Takada A, Kawaoka Y (2003). "Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications". Reviews in Medical Virology. 13 (6): 387–98. doi:10.1002/rmv.405. PMID 14625886.
- Takada A, Feldmann H, Ksiazek TG, Kawaoka Y (July 2003). "Antibody-dependent enhancement of Ebola virus infection". Journal of Virology. 77 (13): 7539–44. doi:10.1128/JVI.77.13.7539-7544.2003. PMC 164833. PMID 12805454.