Viral metagenomics
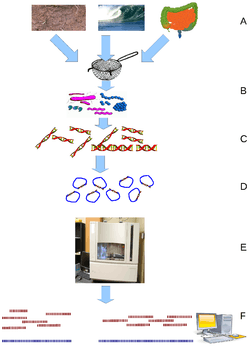
Viral metagenomics is the study of viral genetic material sourced directly from the environment rather than from a host or natural reservoir. The goal is to ascertain viral diversity in the environment that is often missed in studies targeting specific potential reservoirs. It reveals important information on virus evolution and the genetic diversity of the viral community without the need for isolating viral species and cultivating them in the laboratory. Some of the first metagenomic studies of viruses were done with ocean samples, and revealed that most of the sequences of DNA and RNA viruses had no matches in databases.[1][2] This approach has created improvements in molecular epidemiology and accelerated the discovery of novel viruses.[3][4] Acknowledging the importance of viral metagenomics, the International Committee on Taxonomy of Viruses (ICTV) recognizes that genomes assembled from metagenomic data represent actual viruses and encourages their official classification following the same procedures as those used for viruses isolated and characterized using classical virology approaches.[5] In addition, the IMG/VR system [6] -the largest interactive public virus database with over 700,000 metagenomic viral sequences and isolate viruses, serves as a starting point for the sequence analysis of viral fragments derived from metagenomic samples. The virus detection method and host assignment approach in IMG/VR is described in a paper discussing Earth's virome [7] and is fully presented as a protocol [8].
See also
References
- ↑ Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F (2006). "The marine viromes of four oceanic regions". PLoS Biology. 4: e368. doi:10.1371/journal.pbio.0040368. PMC 1634881. PMID 17090214.
- ↑ Culley, A. I.; Lang, A. S.; Suttle, C. A. (2006). "Metagenomic analysis of coastal RNA virus communities". Science. 312 (5781): 1795–1798. doi:10.1126/science.1127404.
- ↑ David M. Kristensen1, Arcady R. Mushegian1, 2, Valerian V. Dolja3 and Eugene V. Koonin4, New dimensions of the virus world discovered through metagenomics. Trends in Microbiology, Volume 18, Issue 1, 11-19, 25 November 2009.
- ↑ Bernardo P, Albina E, Eloit M, Roumagnac P.. Pathology and viral metagenomics, a recent history.[Article in French].Med Sci (Paris). 2013 May;29(5):501-8.
- ↑ Simmonds P, Adams MJ, Benkő M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AMQ, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM (2017). "Consensus statement: Virus taxonomy in the age of metagenomics" (PDF). Nature Reviews Microbiology. 15: 161–168. doi:10.1038/nrmicro.2016.177. PMID 28134265.
- ↑ Paez-Espino D, Chen AI, Palaniappan K, Ratner A, Chu K, Szeto E, Pillay M, Huang J, Markowitz VM, Nielsen T, Huntemann M, Reddy TBK, Pavlopoulos GA, Sullivan MB, Campbell BJ, Chen F, McMahon K, Hallam SJ, Denef V, Cavicchioli R, Caffrey SM, Streit WR, Webster J, Handley KM, Salekdeh GH, Tsesmetzis N, Setubal JC, Pope PB, Liu W, Rivers AR, Ivanova NN, Kyrpides NC (2016). "IMG/VR: A database of cultured and uncultured DNA Viruses and Retroviruses". Nucleic Acids Research. 45: D457–D465. doi:10.1093/nar/gkw1030. PMC 5210529. PMID 27799466.
- ↑ Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC (2016). "Uncovering Earth's Virome". Nature. 536: 425–30. doi:10.1038/nature19094. PMID 27533034.
- ↑ Paez-Espino D, Pavlopoulos GA, Ivanova NN, Kyrpides NC (2016). "Non-targeted virus sequence discovery pipeline and virus clustering for metagenomic data". Nature Protocols. 12: 1673–1682. doi:10.1038/nprot.2017.063. PMID 28749930.
External links
- IMG/VR The IMG Viral Database (IMG/VR).
- CAMERA Cyberinfrastructure for Metagenomics, data repository and tools for metagenomics research.
- GOLD Genomes OnLine Database (GOLD).
- IMG/M The Integrated Microbial Genomes system, for metagenome analysis by the DOE-JGI.
- MEGAN MEtaGenome ANalyzer. A stand-alone metagenome analysis tool.
- MetaGeneMark MetaGeneMark for MetaGenome Gene Finding
- Metagenomics and Our Microbial Planet A website on metagenomics and the vital role of microbes on Earth from the National Academies.
- Metagenomics at the European Bioinformatics Institute Analysis and archiving of metagenomic data.
- Metagenomics: Sequences from the Environment free ebook from NCBI Bookshelf.
- MG-RAST Metagenomics Rapid Annotation using Subsystem Technology server
- The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet A report released by the National Research Council in March 2007. Also, see the Report In Brief.