TSEN15
tRNA-splicing endonuclease subunit Sen15 is an enzyme that in humans is encoded by the TSEN15 gene.[5][6][7]
tRNA splicing is a fundamental process for cell growth and division. SEN15 is a subunit of the tRNA splicing endonuclease, which catalyzes the removal of introns, the first step in tRNA splicing (Paushkin et al., 2004).[supplied by OMIM][7]
Interactions
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000198860 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000014980 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Sood R, Bonner TI, Makalowska I, Stephan DA, Robbins CM, Connors TD, Morgenbesser SD, Su K, Faruque MU, Pinkett H, Graham C, Baxevanis AD, Klinger KW, Landes GM, Trent JM, Carpten JD (Apr 2001). "Cloning and characterization of 13 novel transcripts and the human RGS8 gene from the 1q25 region encompassing the hereditary prostate cancer (HPC1) locus". Genomics. 73 (2): 211–22. doi:10.1006/geno.2001.6500. PMID 11318611.
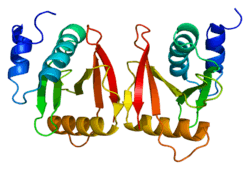
- ↑ Song J, Markley JL (Jan 2007). "Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold". J Mol Biol. 366 (1): 155–64. doi:10.1016/j.jmb.2006.11.024. PMC 1865571. PMID 17166513.
- 1 2 "Entrez Gene: C1orf19 chromosome 1 open reading frame 19".
- ↑ Paushkin, Sergey V; Patel Meenal; Furia Bansri S; Peltz Stuart W; Trotta Christopher R (Apr 2004). "Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3' end formation". Cell. United States. 117 (3): 311–21. doi:10.1016/S0092-8674(04)00342-3. ISSN 0092-8674. PMID 15109492.
Further reading
- Rual JF, Venkatesan K, Hao T, et al. (2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Colland F, Jacq X, Trouplin V, et al. (2004). "Functional proteomics mapping of a human signaling pathway". Genome Res. 14 (7): 1324–32. doi:10.1101/gr.2334104. PMC 442148. PMID 15231748.
- Lehner B, Sanderson CM (2004). "A protein interaction framework for human mRNA degradation". Genome Res. 14 (7): 1315–23. doi:10.1101/gr.2122004. PMC 442147. PMID 15231747.
- Paushkin SV, Patel M, Furia BS, et al. (2004). "Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3' end formation". Cell. 117 (3): 311–21. doi:10.1016/S0092-8674(04)00342-3. PMID 15109492.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.