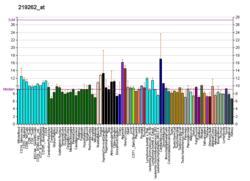
SUV39H2
Histone-lysine N-methyltransferase SUV39H2 is an enzyme that in humans is encoded by the SUV39H2 gene.[5][6]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000152455 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000026646 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, Sattler L, Mattei MG, Denny P, Brown SD, Schweizer D, Jenuwein T (Dec 2000). "Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression". Mol Cell Biol. 20 (24): 9423–33. doi:10.1128/MCB.20.24.9423-9433.2000. PMC 102198. PMID 11094092.
- ↑ "Entrez Gene: SUV39H2 suppressor of variegation 3-9 homolog 2 (Drosophila)".
Further reading
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, et al. (1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Rea S, Eisenhaber F, O'Carroll D, et al. (2000). "Regulation of chromatin structure by site-specific histone H3 methyltransferases". Nature. 406 (6796): 593–9. doi:10.1038/35020506. PMID 10949293.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Ait-Si-Ali S, Guasconi V, Fritsch L, et al. (2005). "A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells". EMBO J. 23 (3): 605–15. doi:10.1038/sj.emboj.7600074. PMC 1271807. PMID 14765126.
- Frontelo P, Leader JE, Yoo N, et al. (2004). "Suv39h histone methyltransferases interact with Smads and cooperate in BMP-induced repression". Oncogene. 23 (30): 5242–51. doi:10.1038/sj.onc.1207660. PMID 15107829.
- Deloukas P, Earthrowl ME, Grafham DV, et al. (2004). "The DNA sequence and comparative analysis of human chromosome 10". Nature. 429 (6990): 375–81. doi:10.1038/nature02462. PMID 15164054.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Rual JF, Venkatesan K, Hao T, et al. (2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Yoon KA, Hwangbo B, Kim IJ, et al. (2006). "Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer". Carcinogenesis. 27 (11): 2217–22. doi:10.1093/carcin/bgl084. PMID 16774942.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.