Ribosomal pause
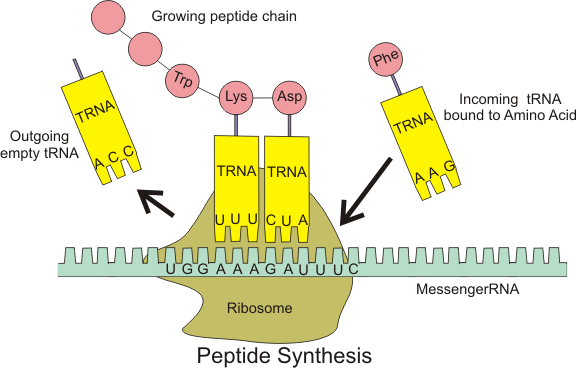
Ribosomal pause refers to the queueing or stacking of ribosomes during translation of the nucleotide sequence of mRNA transcripts. These transcripts are decoded and converted into amino acid sequence during protein synthesis by ribosomes. Within the ribosome, transfer RNA molecules recognise specific trinucleotide codons on the mRNA, and add their cognate amino acids to nascent protein chains.[1][2]
It’s been known since the 1980s that different mRNAs are translated at different rates. The main reason for these differences was thought to be the concentration of rarer varieties of tRNA limiting the rate at which some transcripts could be decoded.[3]
Two techniques can localize the ribosomal pause site in vivo; a micrococcal nuclease protection assay and isolation of polysomal transcript, by taking the tissue that is extracted through a sucrose cushion including the translation elongation inhabit it and centrifuge it.[4]
Effects on gene expression
During protein synthesis, rapidly changing conditions in the cell can cause ribosomal pausing. In bacteria and viruses, this can affect growth rate and trigger translational abandonment. This releases the ribosome from the mRNA and the incomplete polypeptide is targeted for destruction.[5]
In eukaryotes, ribosomal pausing can initiate an analogous process which triggers endonucleolytic attack of the mRNA, a process termed mRNA no-go decay. Ribosomal pausing also aids co-translational folding of the nascent polypeptide on the ribosome, and delays protein translation while its encoding mRNA. This can trigger ribosomal frameshifting.[5]
See also
References
- ↑ Jha SS, Komar AA (July 2012). "Isolation of ribosome bound nascent polypeptides in vitro to identify translational pause sites along mRNA". Journal of Visualized Experiments (65). doi:10.3791/4026. PMC 3471273. PMID 22806127.
- ↑ Lopinski JD, Dinman JD, Bruenn JA (February 2000). "Kinetics of ribosomal pausing during programmed -1 translational frameshifting". Molecular and Cellular Biology. 20 (4): 1095–103. doi:10.1128/MCB.20.4.1095-1103.2000. PMC 85227. PMID 10648594.
- ↑ Kontos H, Napthine S, Brierley I (December 2001). "Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency". Molecular and Cellular Biology. 21 (24): 8657–70. doi:10.1128/MCB.21.24.8657-8670.2001. PMC 100026. PMID 11713298.
- ↑ Kim, Jeong-Kook; Hollingsworth, Margaret J. (October 1992). "Localization of in vivo ribosome pause sites". Analytical Biochemistry. 206 (1): 183–188. doi:10.1016/s0003-2697(05)80031-4. ISSN 0003-2697.
- 1 2 Buchan JR, Stansfield I (September 2007). "Halting a cellular production line: responses to ribosomal pausing during translation". Biology of the Cell. 99 (9): 475–87. doi:10.1042/BC20070037. PMID 17696878.